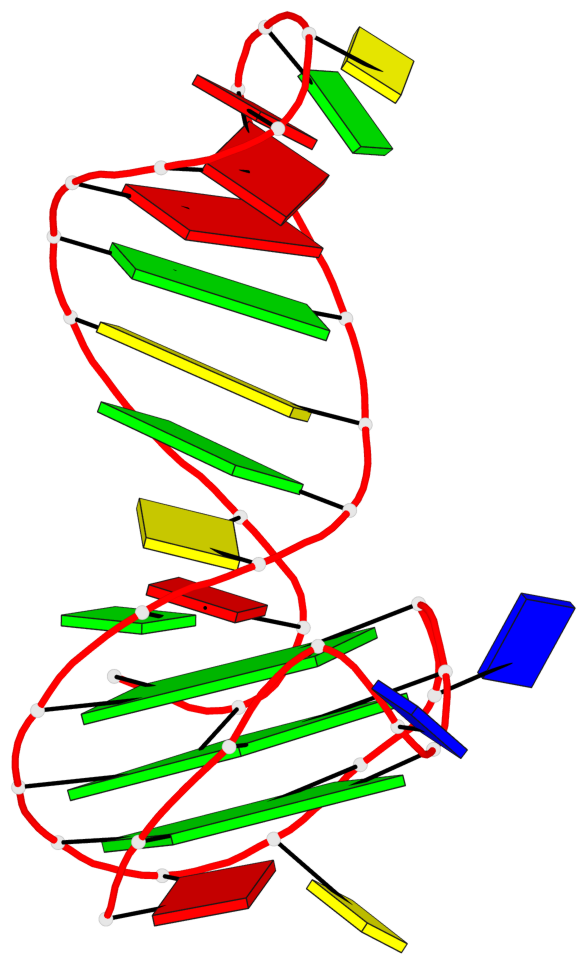
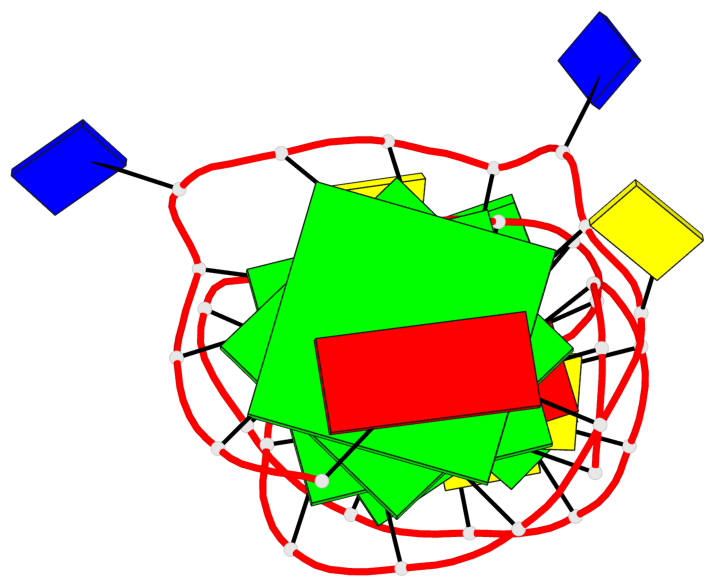
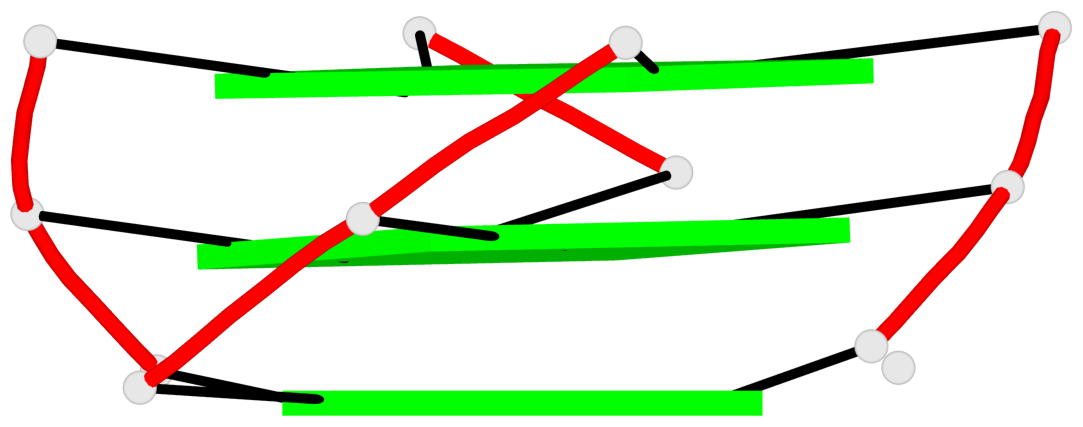
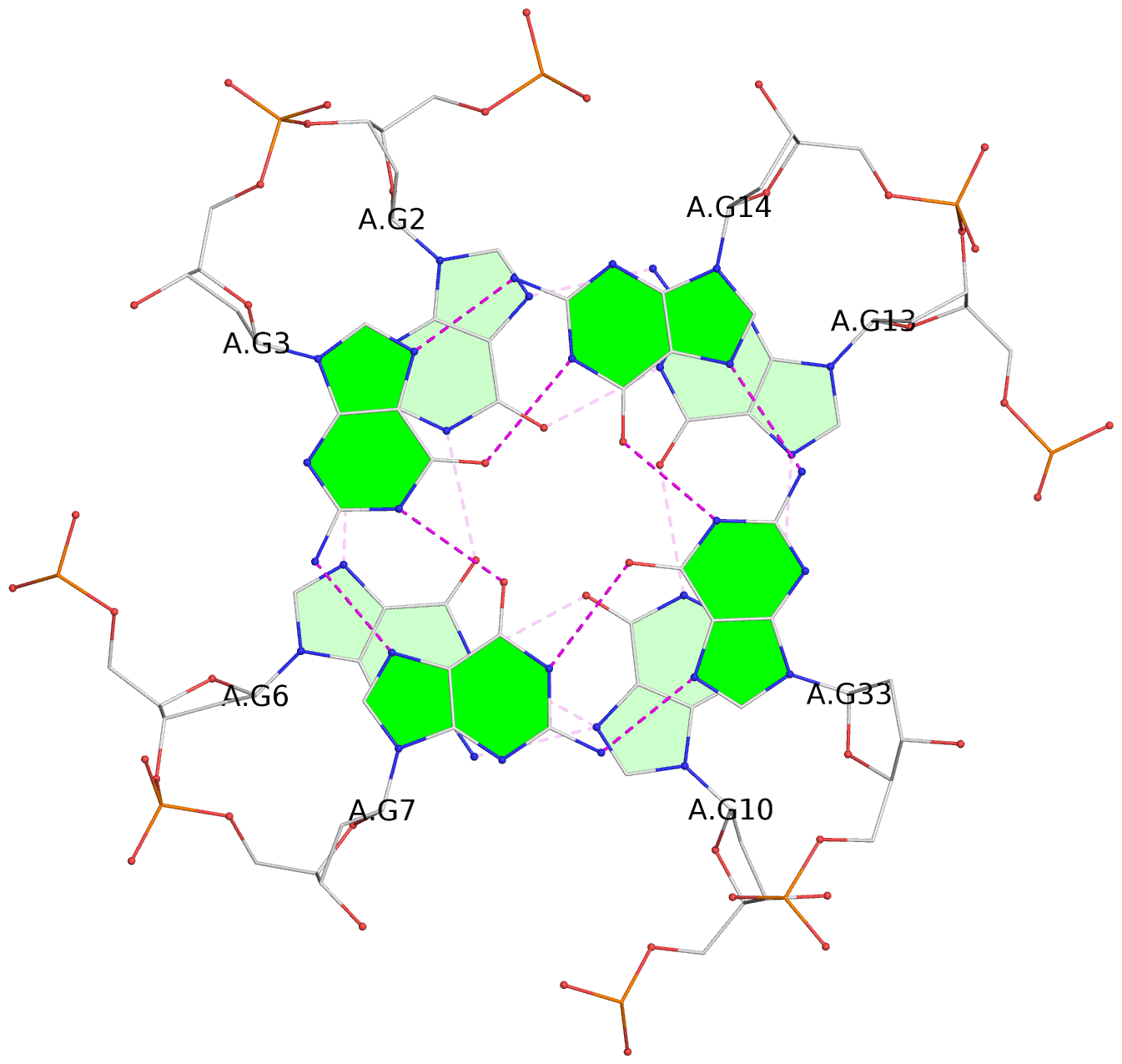
Detailed DSSR results for the G-quadruplex: PDB entry 2m92
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2m92
- Class
- DNA
- Method
- NMR
- Summary
- Structure of d[agggtgggtgctggggcgcgaagcattcgcgagg] quadruplex-duplex hybrid
- Reference
- Lim KW, Phan AT (2013): "Structural basis of DNA quadruplex-duplex junction formation." Angew.Chem.Int.Ed.Engl., 52, 8566-8569. doi: 10.1002/anie.201302995.
- Abstract
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 2(-PX+P), parallel(4+0), UUUU
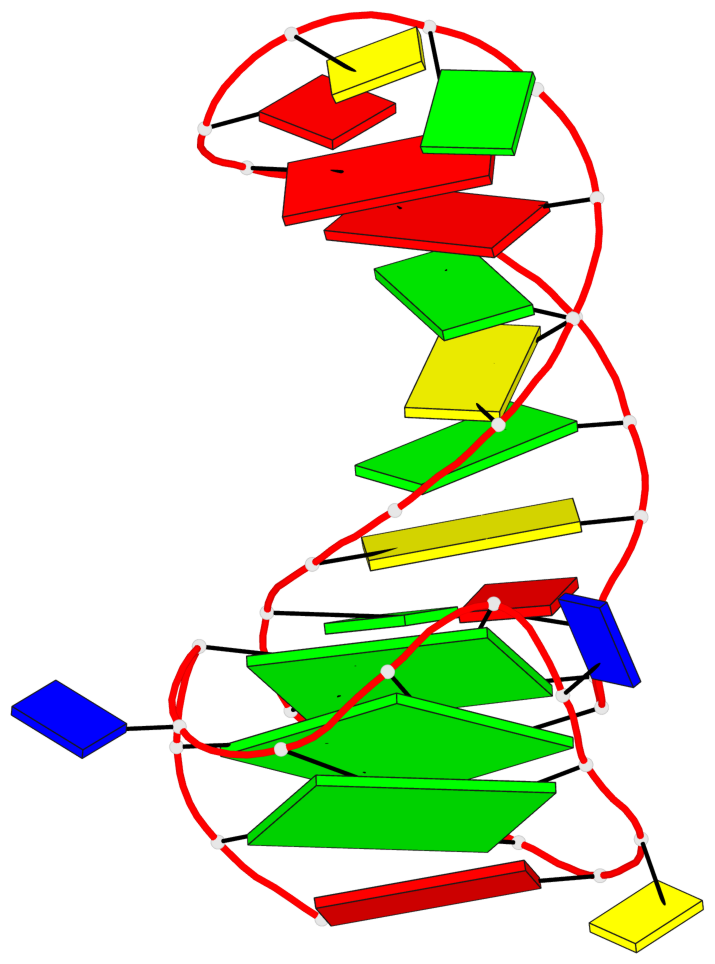
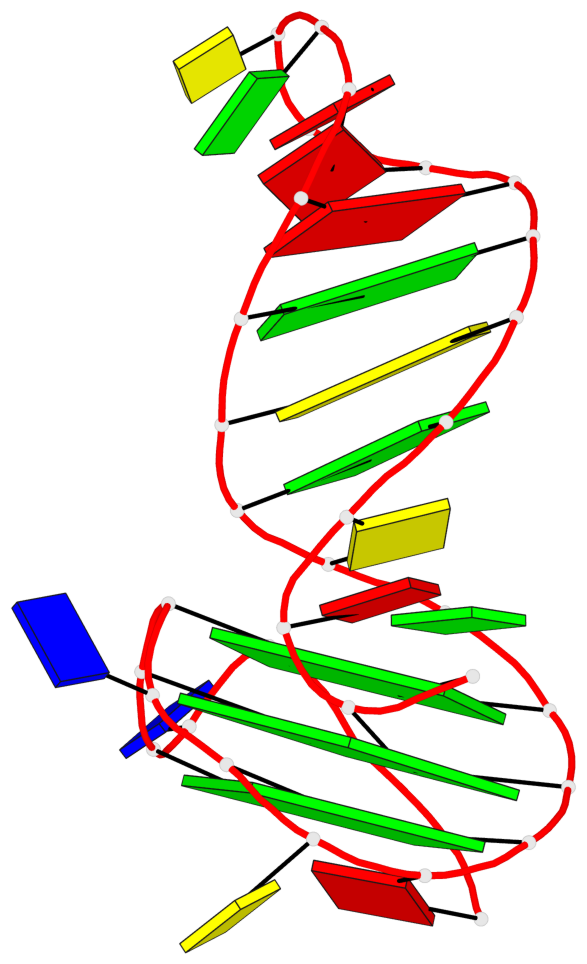
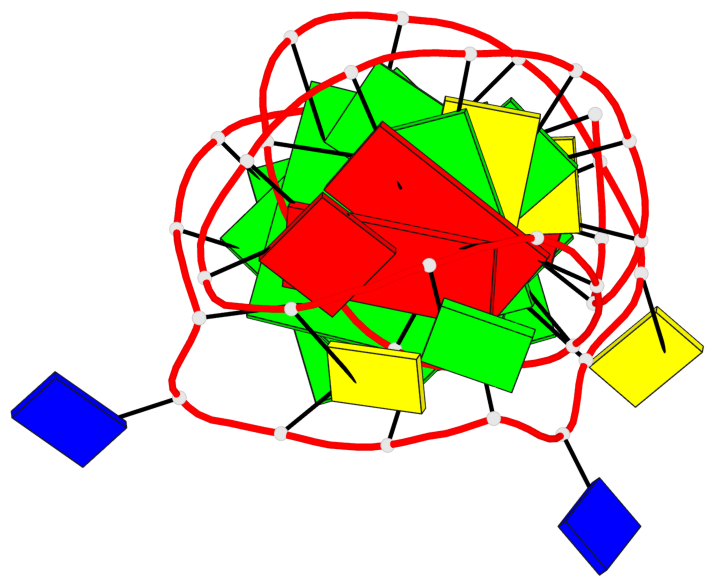
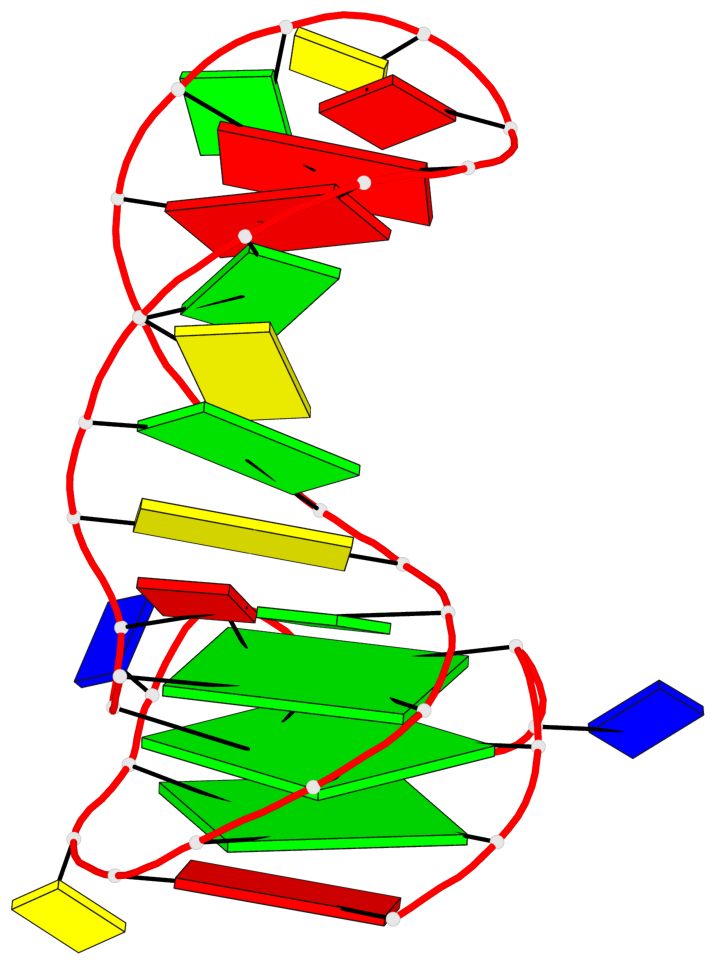
Base-block schematics in six views
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.068 type=planar nts=4 GGGG A.DG2,A.DG6,A.DG10,A.DG13 2 glyco-bond=---- sugar=--3- groove=---- planarity=0.141 type=planar nts=4 GGGG A.DG3,A.DG7,A.DG33,A.DG14 3 glyco-bond=---- sugar=---- groove=---- planarity=0.100 type=planar nts=4 GGGG A.DG4,A.DG8,A.DG34,A.DG15
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-PX+P), parallel(4+0)
List of 2 non-stem G4-loops (including the two closing Gs)
1* type=V-shaped helix=#1 nts=5 GGGTG A.DG6,A.DG7,A.DG8,A.DT9,A.DG10 2 type=lateral helix=#1 nts=4 GCTG A.DG10,A.DC11,A.DT12,A.DG13