Detailed DSSR results for the G-quadruplex: PDB entry 2mb3
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2mb3
- Class
- DNA-DNA inhibitor
- Method
- NMR
- Summary
- Solution structure of an intramolecular (3+1) human telomeric g-quadruplex bound to a telomestatin derivative
- Reference
- Chung WJ, Heddi B, Tera M, Iida K, Nagasawa K, Phan AT (2013): "Solution structure of an intramolecular (3 + 1) human telomeric g-quadruplex bound to a telomestatin derivative." J.Am.Chem.Soc., 135, 13495-13501. doi: 10.1021/ja405843r.
- Abstract
- Guanine-rich human telomeric DNA can adopt secondary structures known as G-quadruplexes, which can be targeted by small molecules to achieve anticancer effects. So far, the structural information on complexes between human telomeric DNA and ligands is limited to the parallel G-quadruplex conformation, despite the high structural polymorphism of human telomeric G-quadruplexes. No structure has been yet resolved for the complex with telomestatin, one of the most promising G-quadruplex-targeting anticancer drug candidates. Here we present the first high-resolution structure of the complex between an intramolecular (3 + 1) human telomeric G-quadruplex and a telomestatin derivative, the macrocyclic hexaoxazole L2H2-6M(2)OTD. This compound is observed to interact with the G-quadruplex through π-stacking and electrostatic interactions. This structural information provides a platform for the design of topology-specific G-quadruplex-targeting compounds and is valuable for the development of new potent anticancer drugs.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-Lw-Ln), hybrid-1(3+1), UUDU
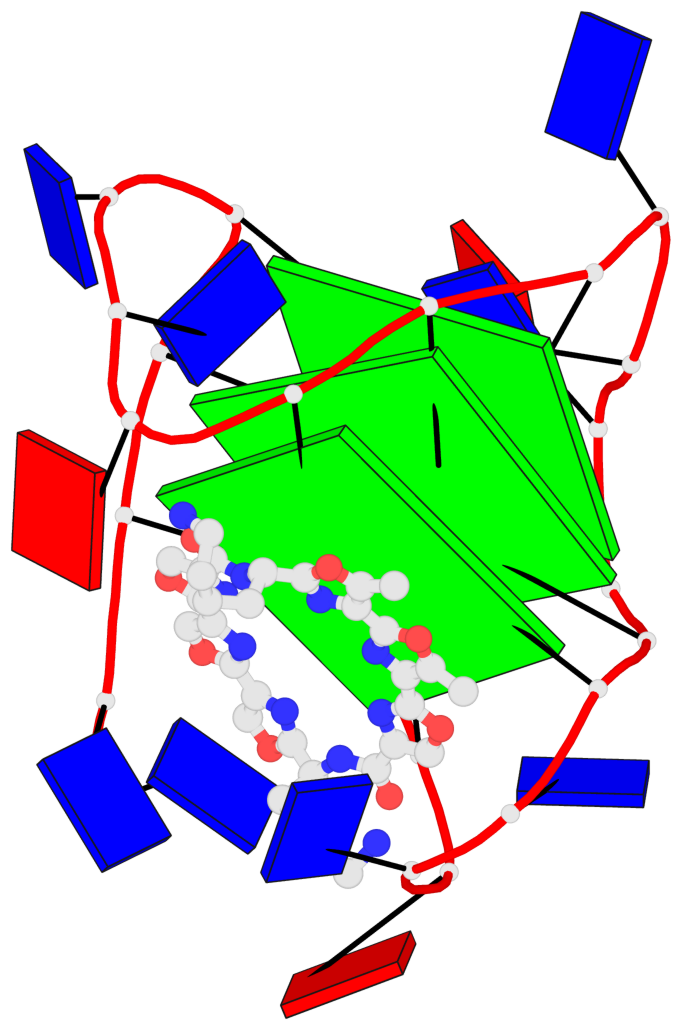
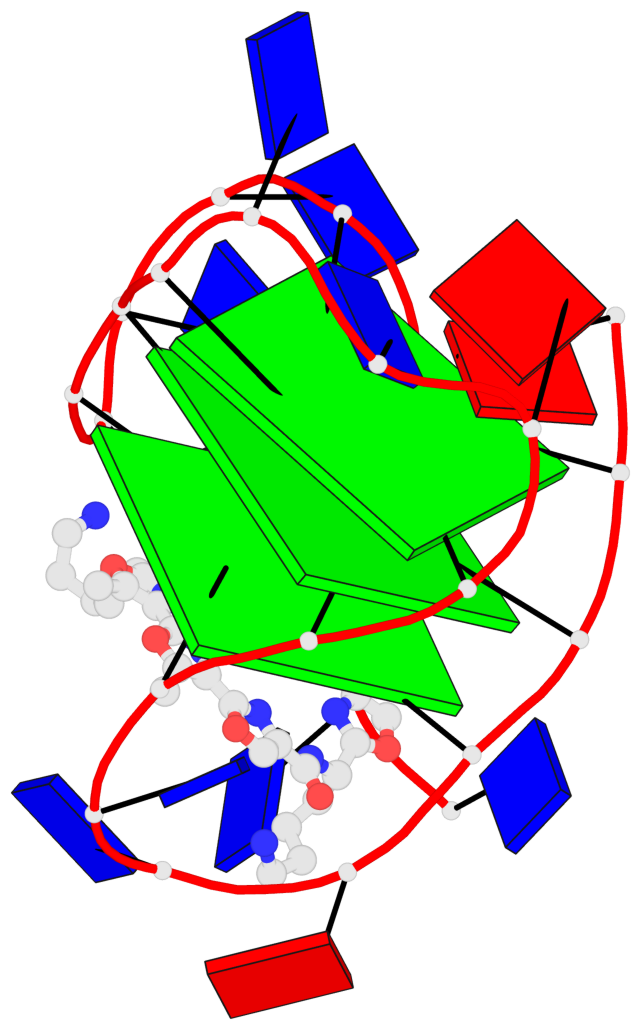
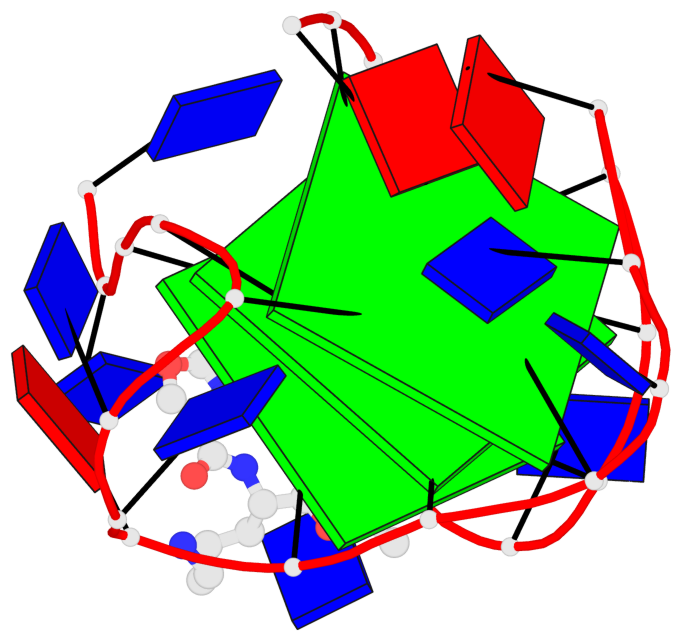
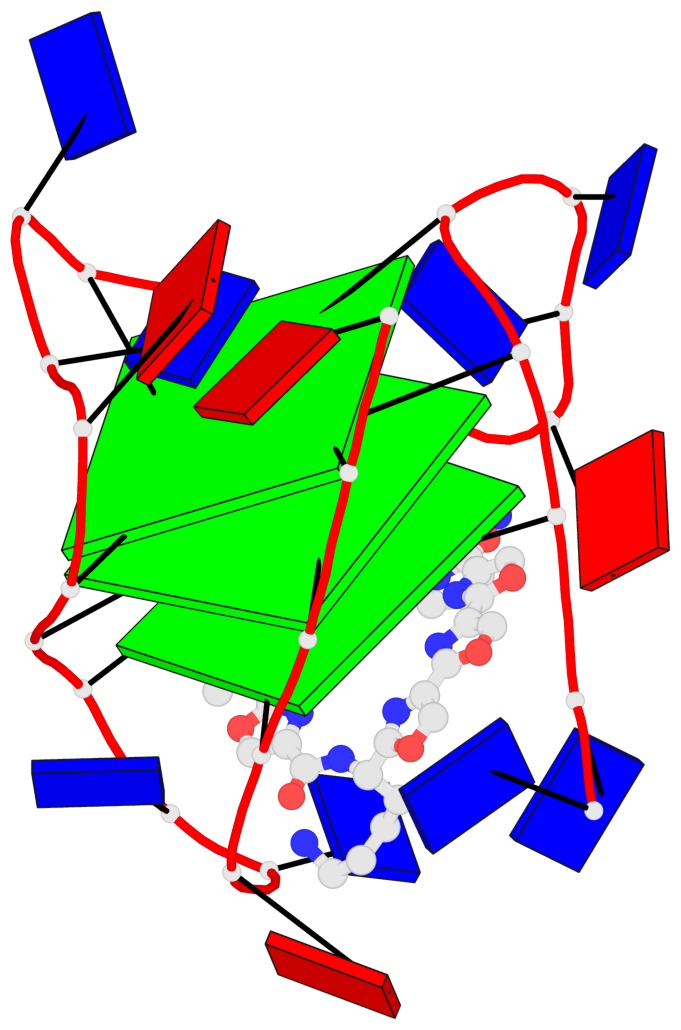
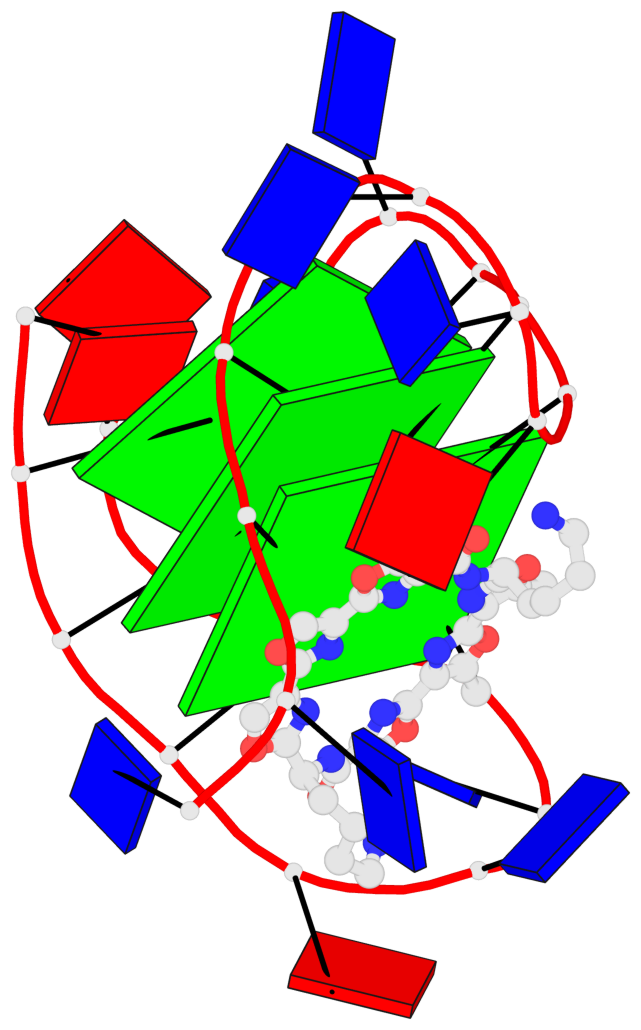
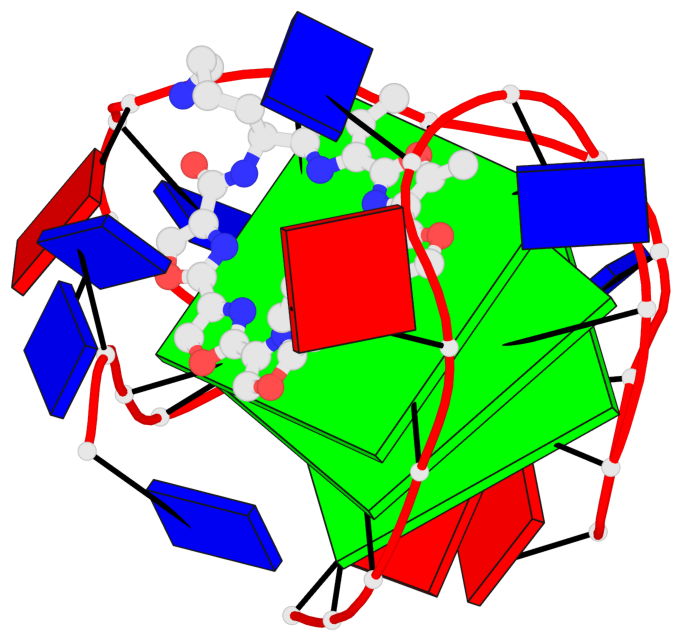
Base-block schematics in six views
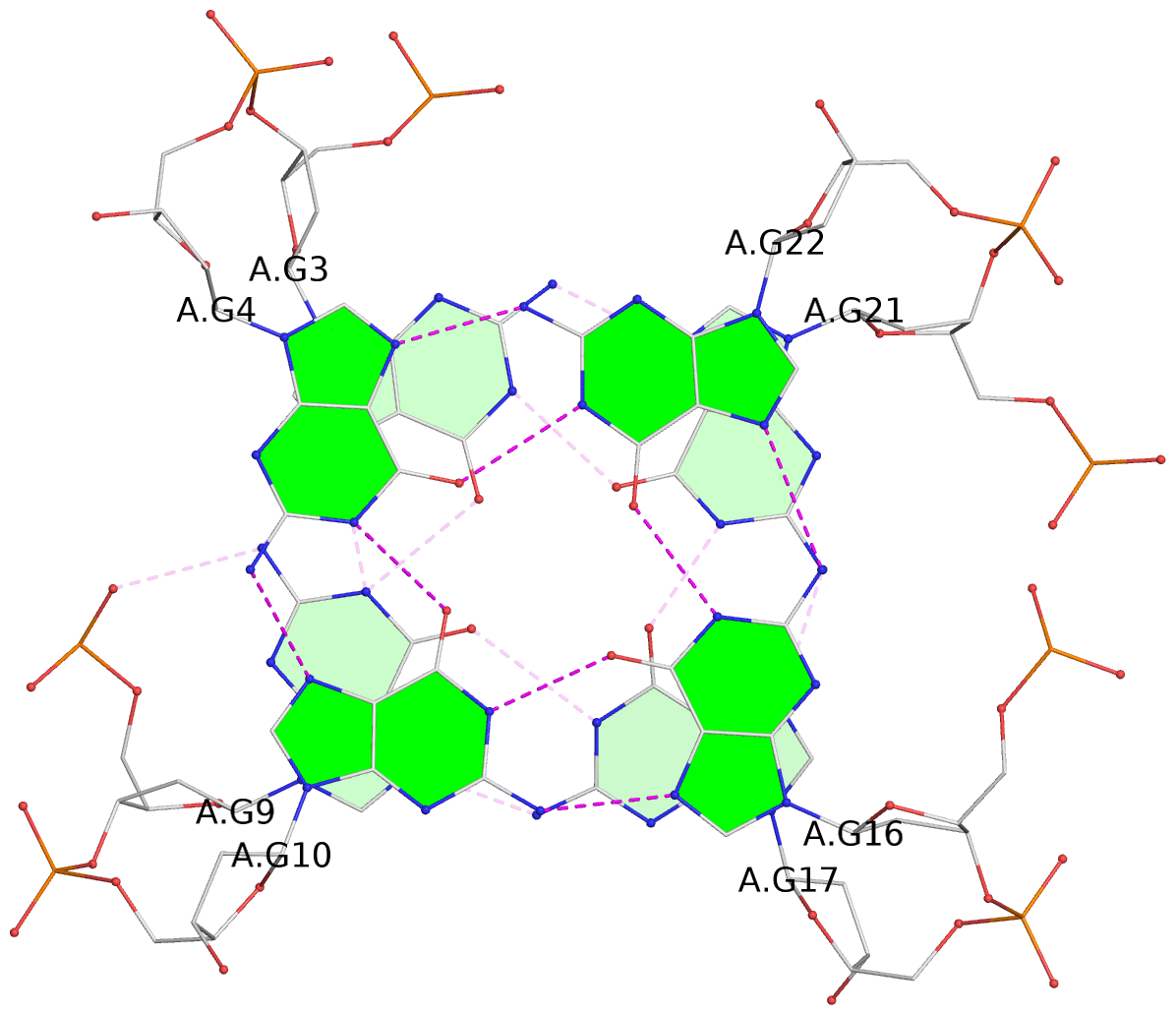
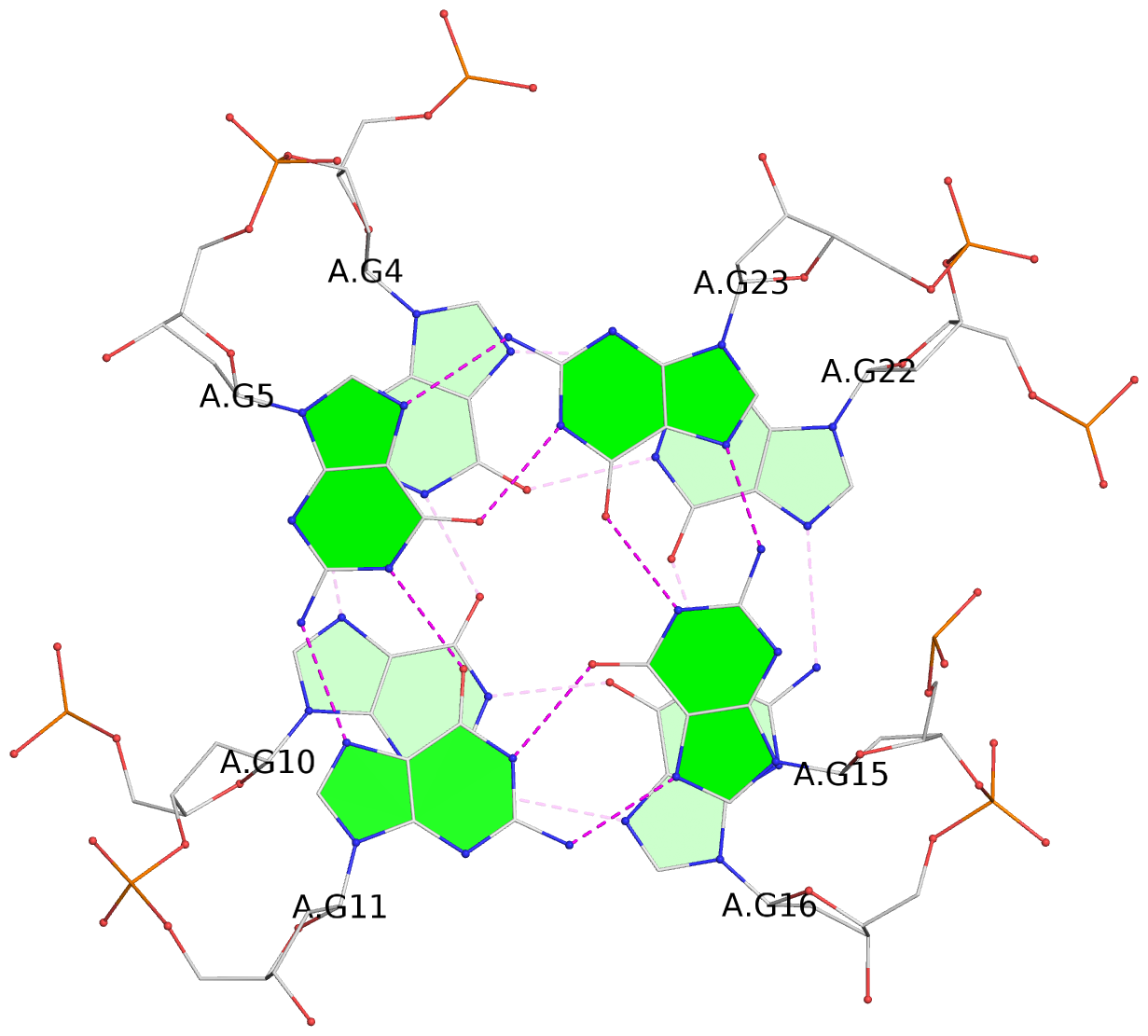
List of 3 G-tetrads
1 glyco-bond=ss-s sugar=.--- groove=-wn- planarity=0.325 type=other nts=4 GGGG A.DG3,A.DG9,A.DG17,A.DG21 2 glyco-bond=--s- sugar=.--- groove=-wn- planarity=0.314 type=other nts=4 GGGG A.DG4,A.DG10,A.DG16,A.DG22 3 glyco-bond=--s- sugar=---- groove=-wn- planarity=0.347 type=other nts=4 GGGG A.DG5,A.DG11,A.DG15,A.DG23
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.