Detailed DSSR results for the G-quadruplex: PDB entry 2mbj
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2mbj
- Class
- DNA
- Method
- NMR
- Summary
- Structure of an antiparallel (2+2) g-quadruplex formed by human telomeric repeats in na+ solution (with g22-to-brg substitution)
- Reference
- Lim KW, Ng VC, Martin-Pintado N, Heddi B, Phan AT (2013): "Structure of the human telomere in Na+ solution: an antiparallel (2+2) G-quadruplex scaffold reveals additional diversity." Nucleic Acids Res., 41, 10556-10562. doi: 10.1093/nar/gkt771.
- Abstract
- Single-stranded DNA overhangs at the ends of human telomeric repeats are capable of adopting four-stranded G-quadruplex structures, which could serve as potential anticancer targets. Out of the five reported intramolecular human telomeric G-quadruplex structures, four were formed in the presence of K(+) ions and only one in the presence of Na(+) ions, leading often to a perception that this structural polymorphism occurs exclusively in the presence of K(+) but not Na(+). Here we present the structure of a new antiparallel (2+2) G-quadruplex formed by a derivative of a 27-nt human telomeric sequence in Na(+) solution, which comprises a novel core arrangement distinct from the known topologies. This structure complements the previously elucidated basket-type human telomeric G-quadruplex to serve as reference structures in Na(+)-containing environment. These structures, together with the coexistence of other conformations in Na(+) solution as observed by nuclear magnetic resonance spectroscopy, establish the polymorphic nature of human telomeric repeats beyond the influence of K(+) ions.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(+Ln+P+Lw), (2+2), UUDD
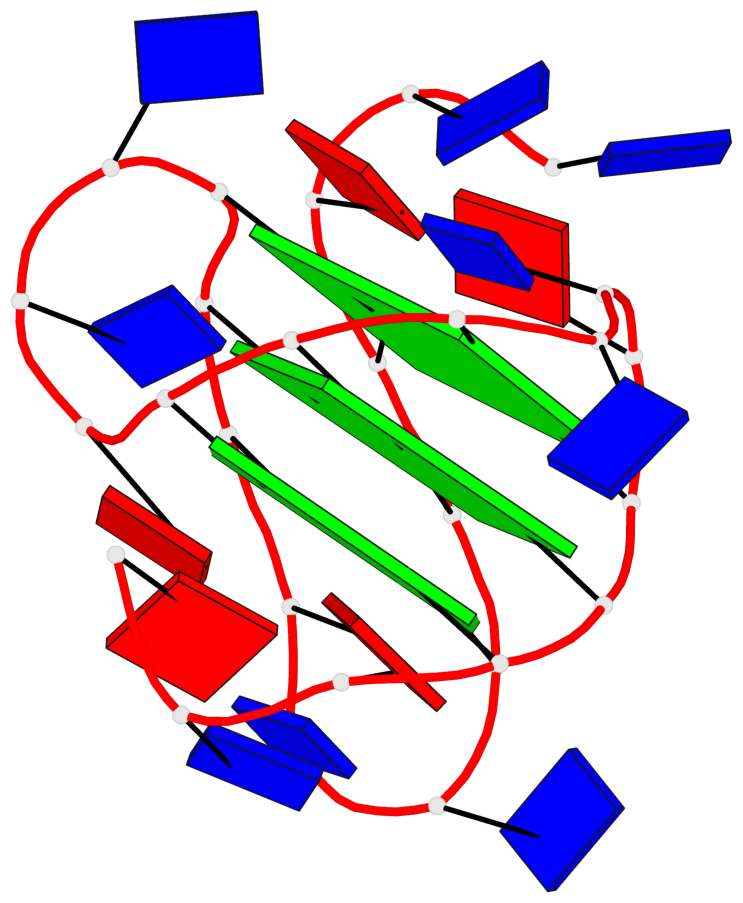
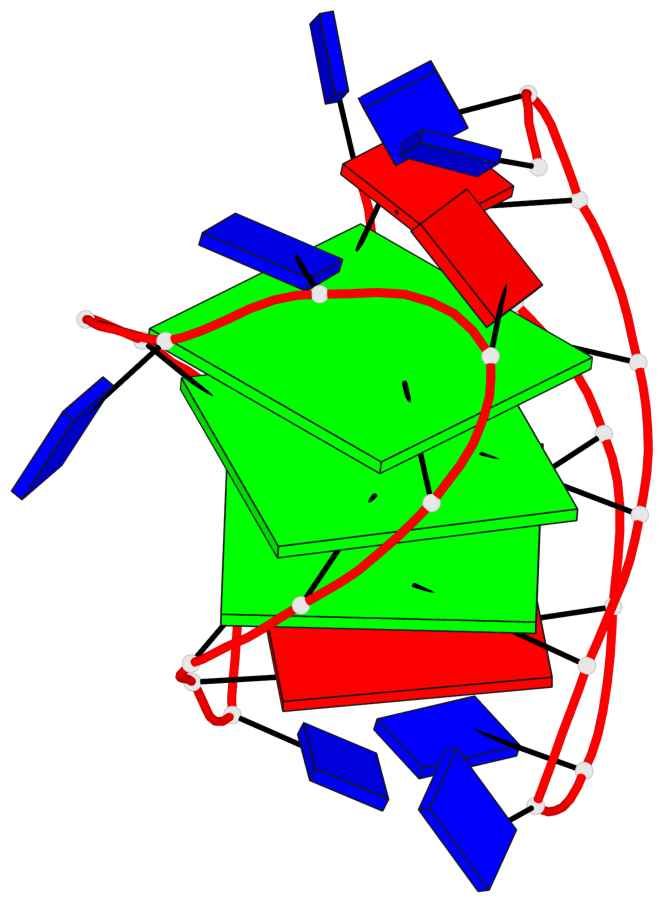
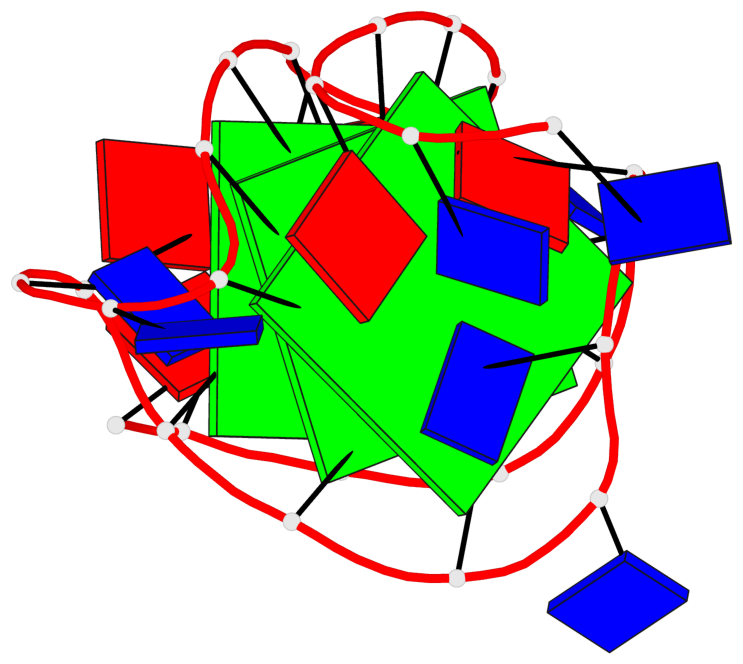
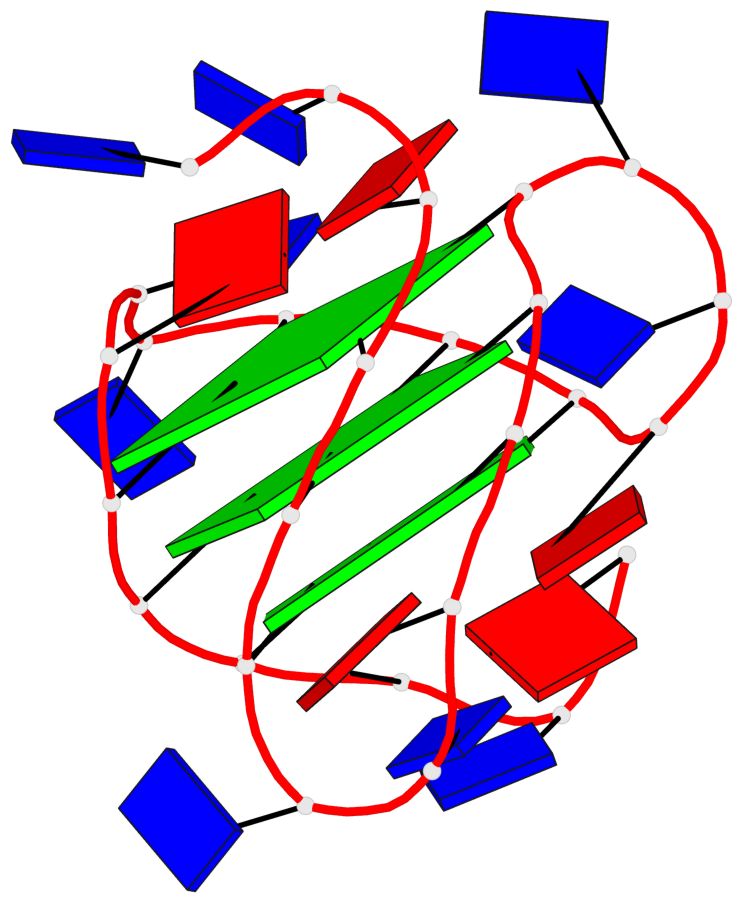
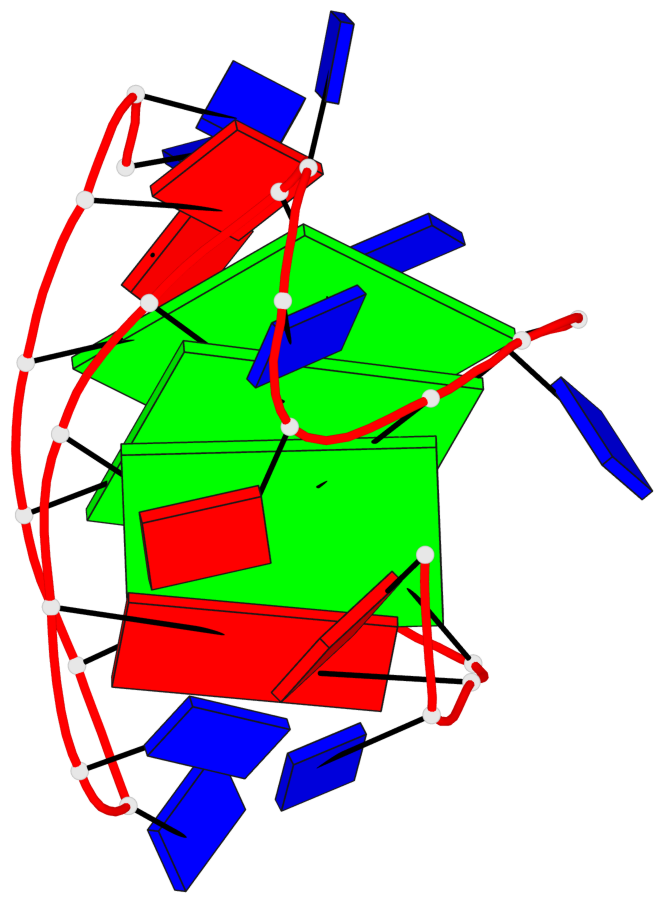
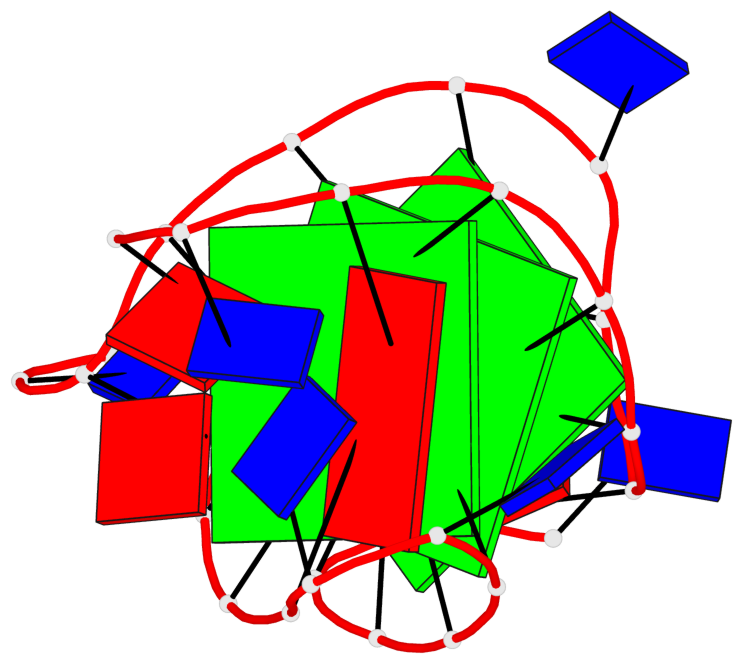
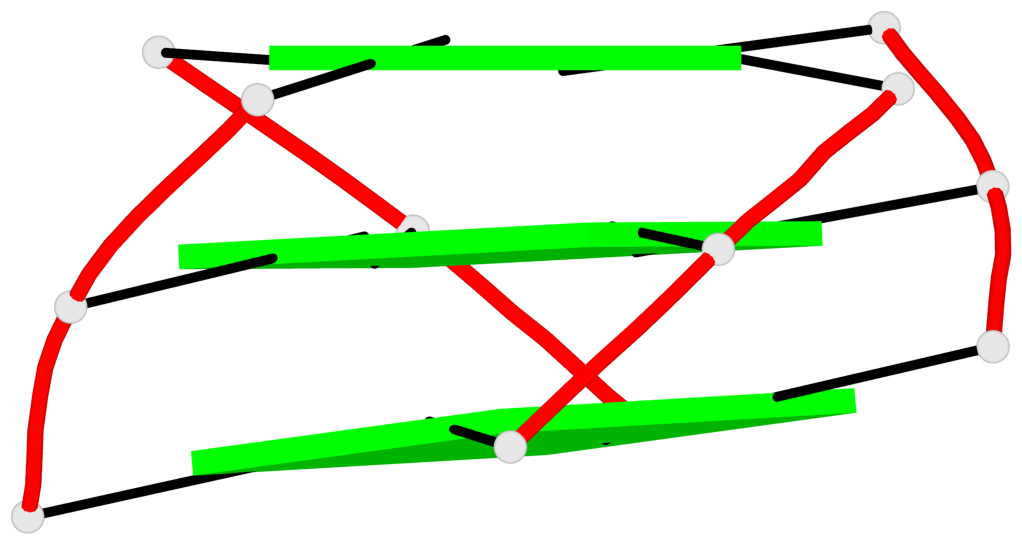
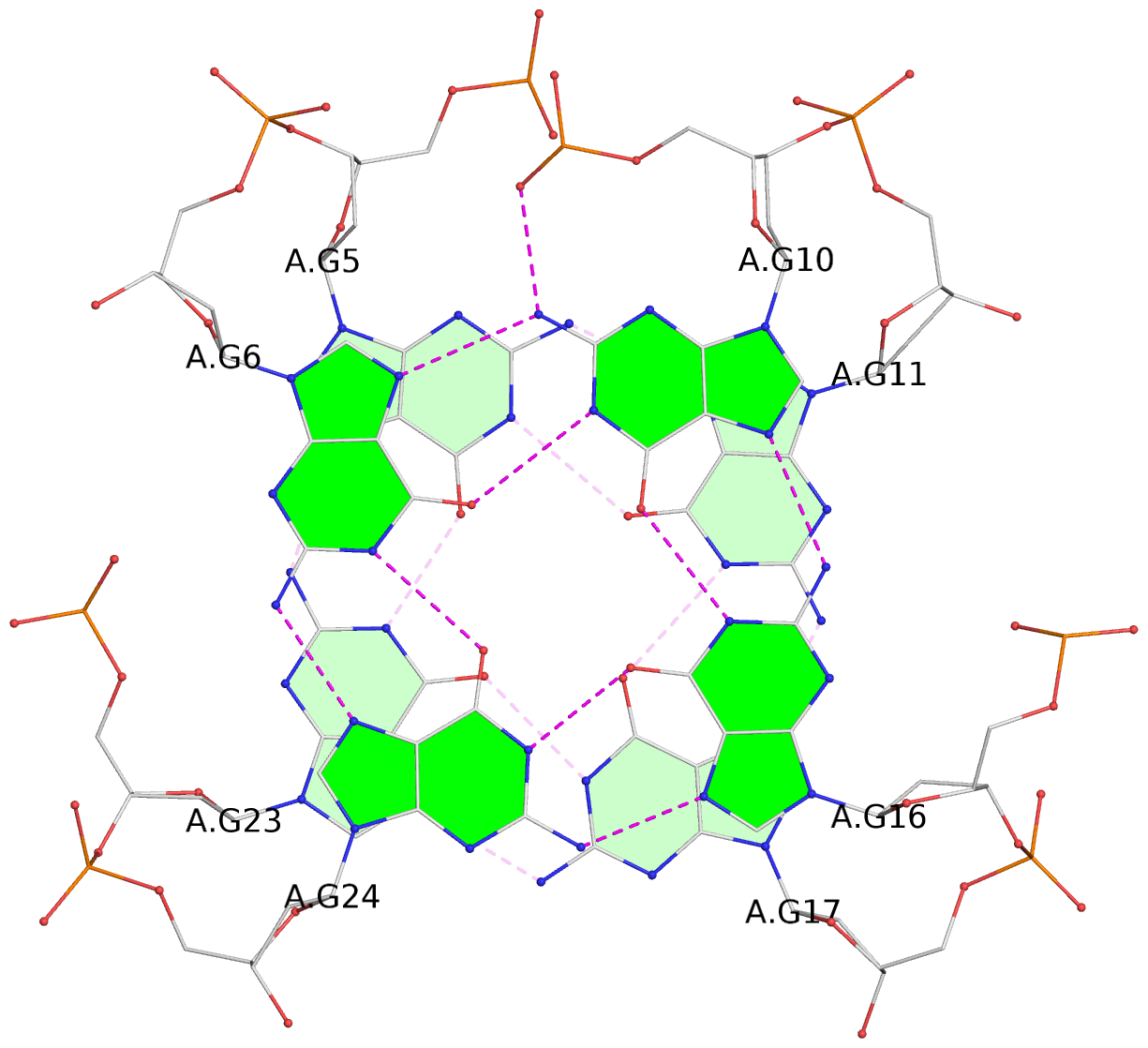
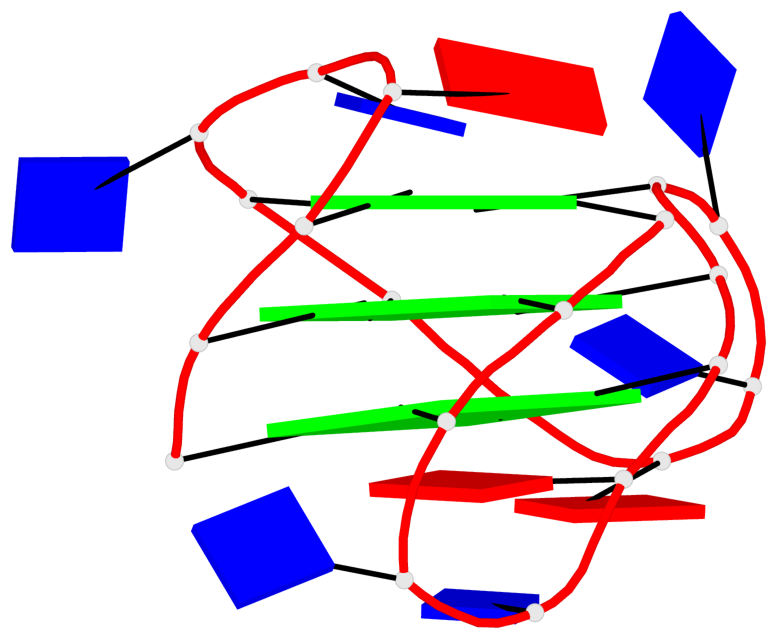
Base-block schematics in six views
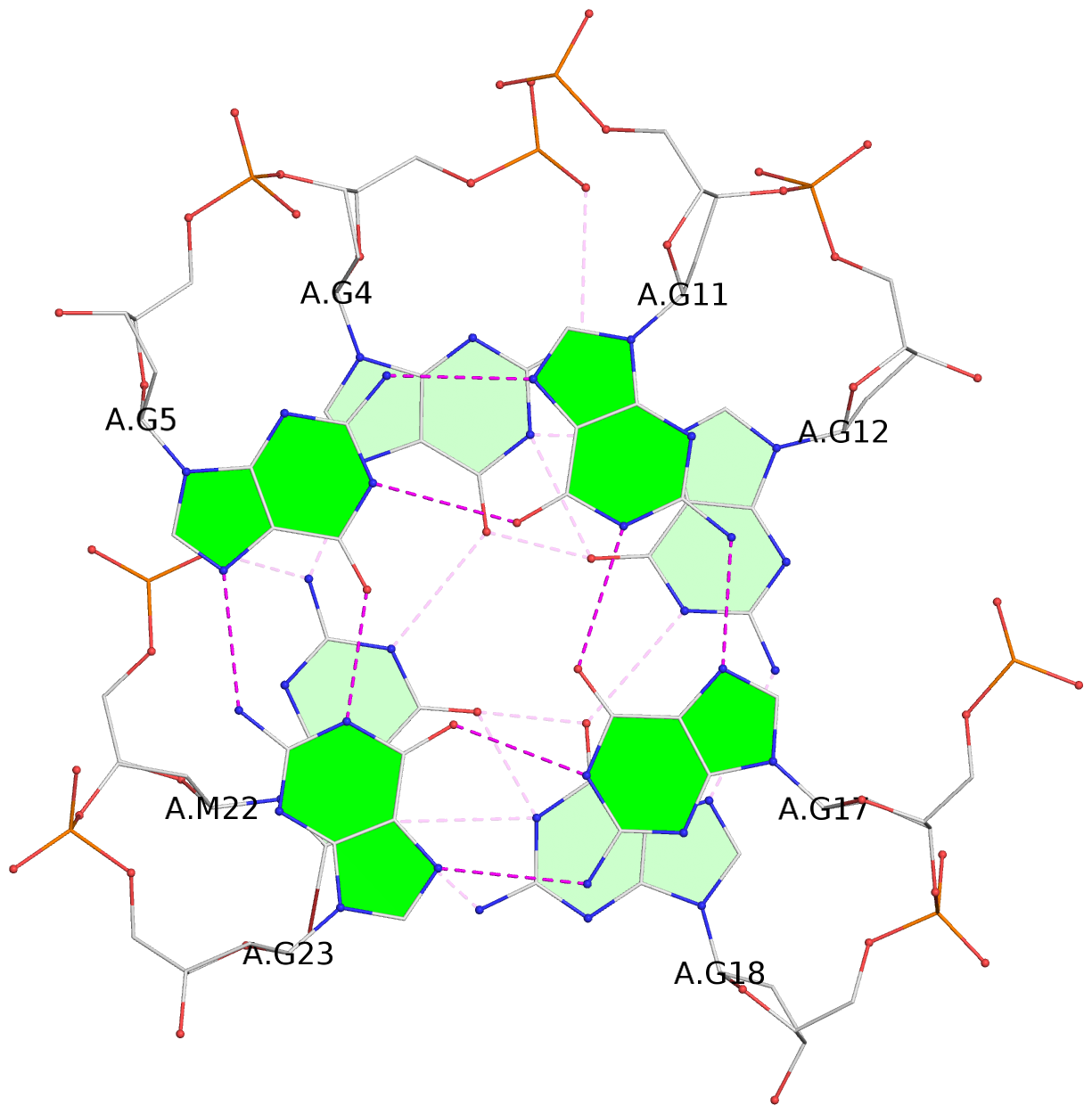
List of 3 G-tetrads
1 glyco-bond=ss-- sugar=---- groove=-w-n planarity=0.121 type=planar nts=4 GgGG A.DG4,A.BGM22,A.DG18,A.DG12 2 glyco-bond=ss-- sugar=---. groove=-w-n planarity=0.115 type=planar nts=4 GGGG A.DG5,A.DG23,A.DG17,A.DG11 3 glyco-bond=--ss sugar=---- groove=-w-n planarity=0.103 type=planar nts=4 GGGG A.DG6,A.DG24,A.DG16,A.DG10
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.