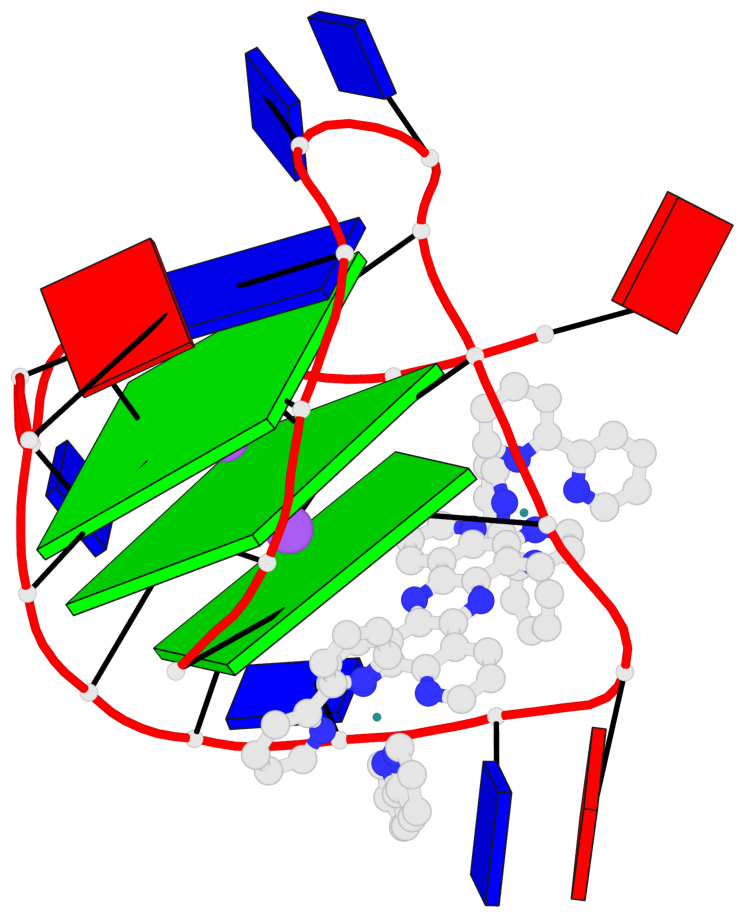
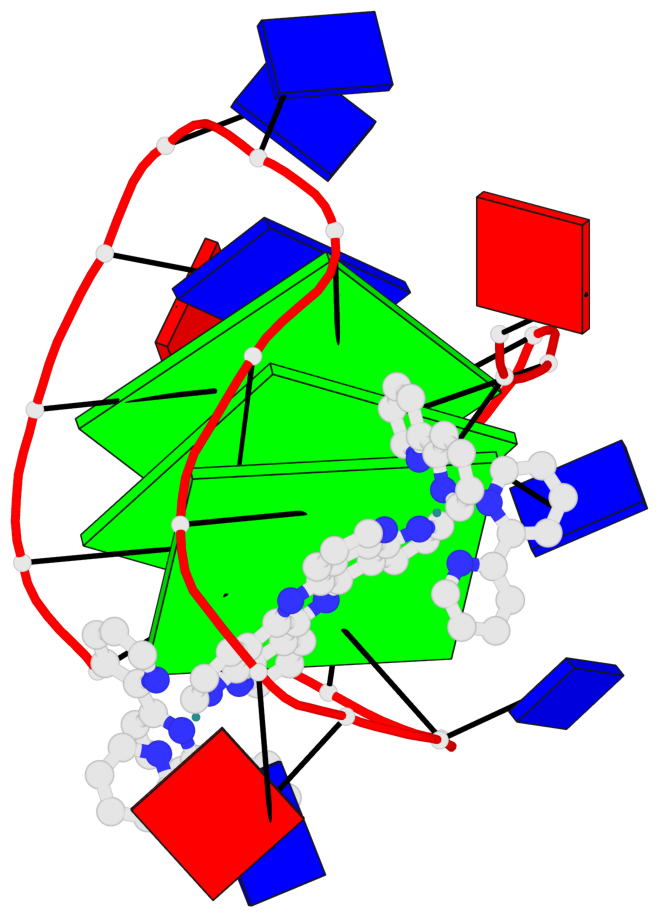
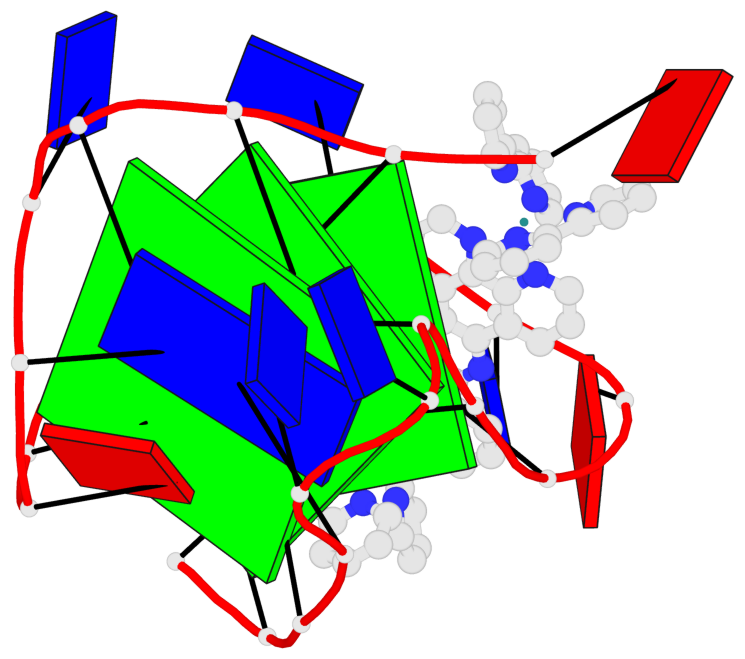
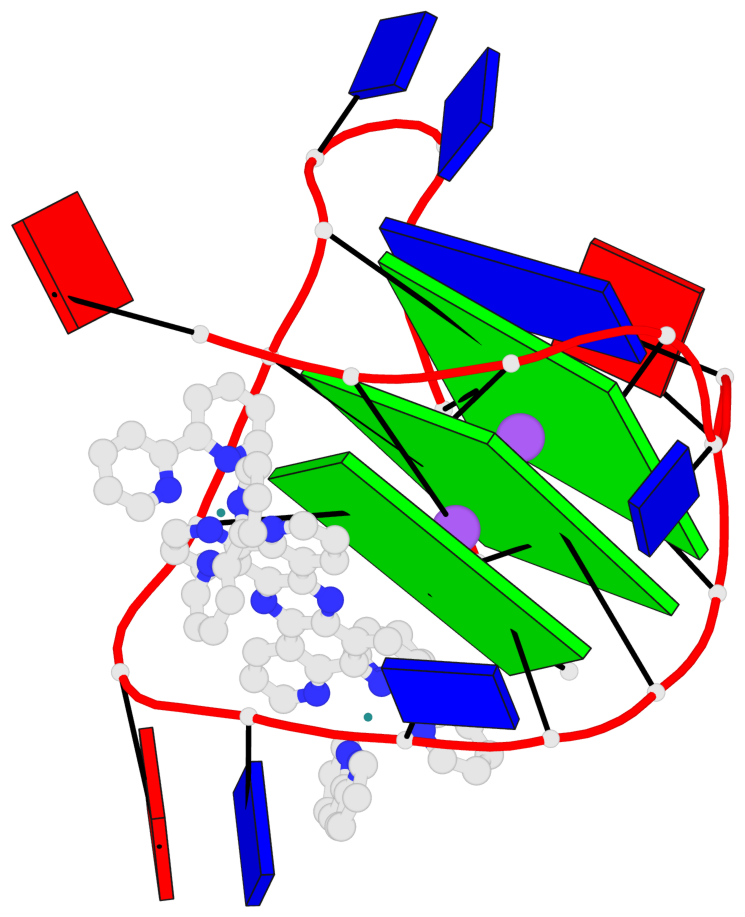
Detailed DSSR results for the G-quadruplex: PDB entry 2mco
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2mco
- Class
- DNA
- Method
- NMR
- Summary
- Structural studies on dinuclear ruthenium(ii) complexes that bind diastereoselectively to an anti-parallel folded human telomere sequence
- Reference
- Wilson T, Costa PJ, Felix V, Williamson MP, Thomas JA (2013): "Structural Studies on Dinuclear Ruthenium(II) Complexes That Bind Diastereoselectively to an Antiparallel Folded Human Telomere Sequence." J.Med.Chem., 56, 8674-8683. doi: 10.1021/jm401119b.
- Abstract
- We report DNA binding studies of the dinuclear ruthenium ligand [{Ru(phen)2}2tpphz](4+) in enantiomerically pure forms. As expected from previous studies of related complexes, both isomers bind with similar affinity to B-DNA and have enhanced luminescence. However, when tested against the G-quadruplex from human telomeres (which we show to form an antiparallel basket structure with a diagonal loop across one end), the ΛΛ isomer binds approximately 40 times more tightly than the ΔΔ, with a stronger luminescence. NMR studies show that the complex binds at both ends of the quadruplex. Modeling studies, based on experimentally derived restraints obtained for the closely related [{Ru(bipy)2}2tpphz](4+), show that the ΛΛ isomer fits neatly under the diagonal loop, whereas the ΔΔ isomer is unable to bind here and binds at the lateral loop end. Molecular dynamics simulations show that the ΔΔ isomer is prevented from binding under the diagonal loop by the rigidity of the loop. We thus present a novel enantioselective binding substrate for antiparallel basket G-quadruplexes, with features that make it a useful tool for quadruplex studies.
- G4 notes
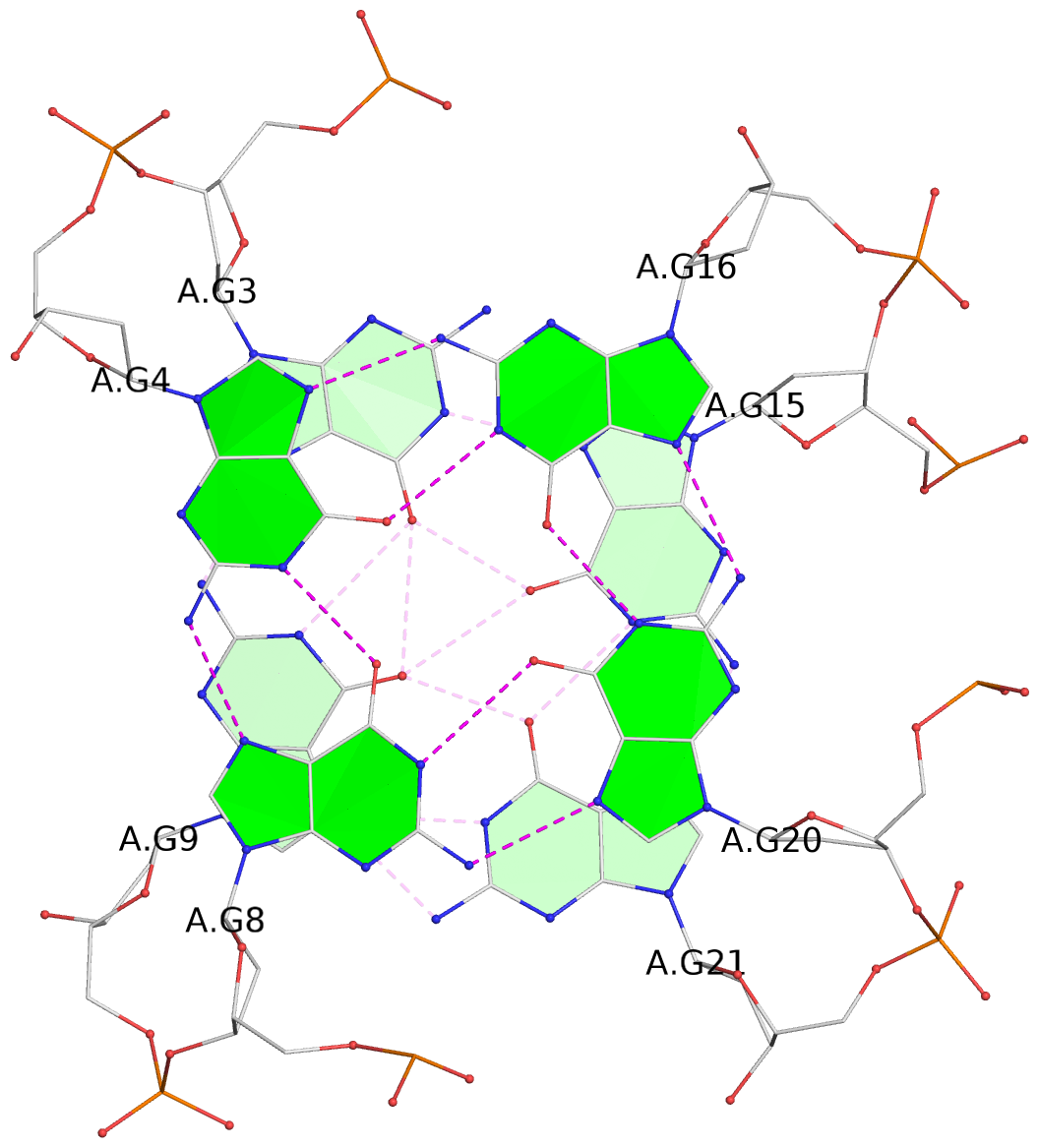
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-LwD+Ln), basket(2+2), UDDU
Base-block schematics in six views
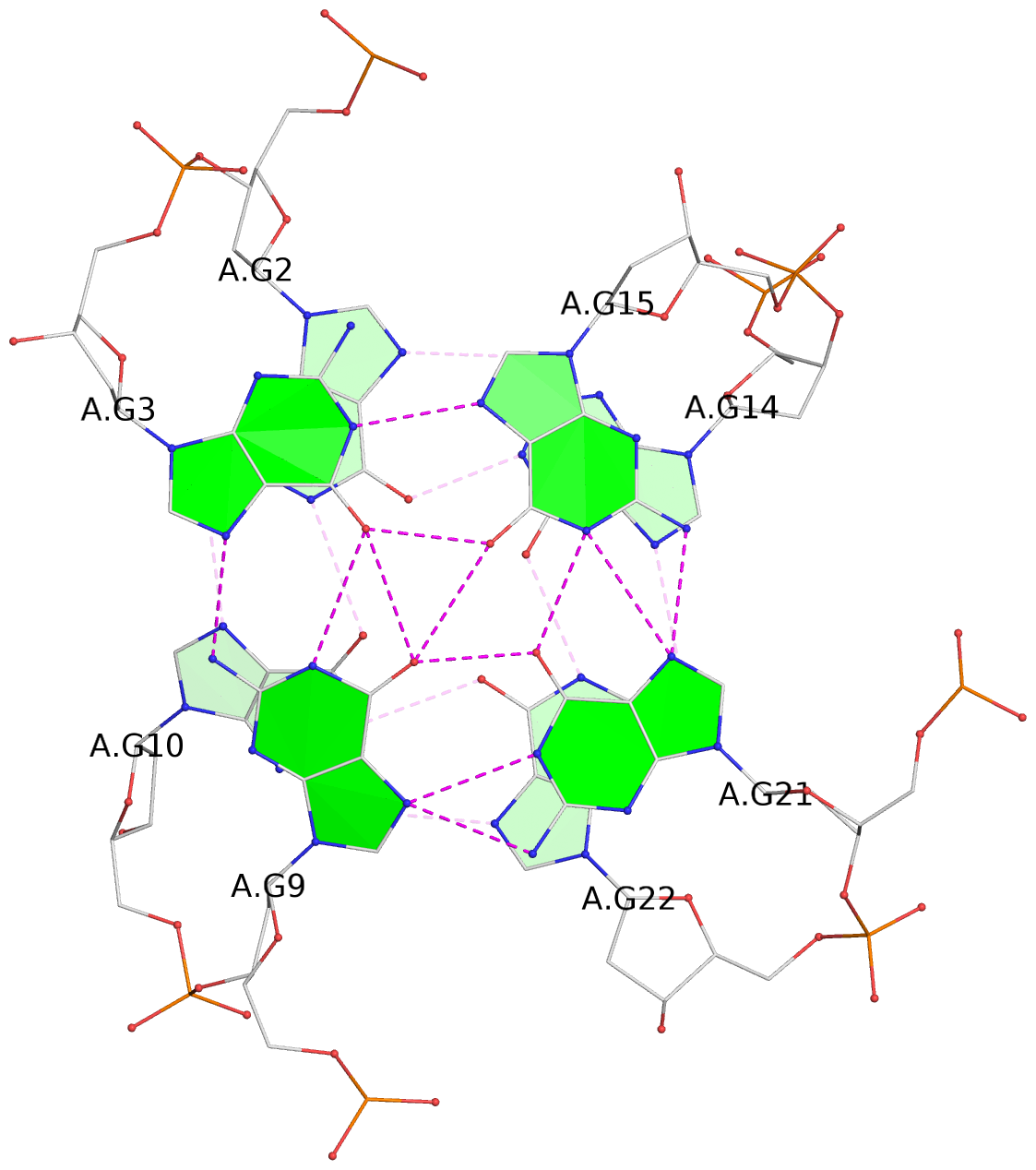
List of 3 G-tetrads
1 glyco-bond=-s-- sugar=..-- groove=wn-- planarity=0.851 type=other nts=4 GGGG A.DG2,A.DG10,A.DG22,A.DG14 2 glyco-bond=s--s sugar=-.-. groove=w-n- planarity=0.542 type=other nts=4 GGGG A.DG3,A.DG9,A.DG21,A.DG15 3 glyco-bond=-ss- sugar=--.- groove=w-n- planarity=0.489 type=saddle nts=4 GGGG A.DG4,A.DG8,A.DG20,A.DG16
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.