Detailed DSSR results for the G-quadruplex: PDB entry 2mwz
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2mwz
- Class
- DNA
- Method
- NMR
- Summary
- Xanthine and 8-oxoguanine in g-quadruplexes: formation of a g g x o tetrad
- Reference
- Cheong VV, Heddi B, Lech CJ, Phan AT (2015): "Xanthine and 8-oxoguanine in G-quadruplexes: formation of a GGXO tetrad." Nucleic Acids Res., 43, 10506-10514. doi: 10.1093/nar/gkv826.
- Abstract
- G-quadruplexes are four-stranded structures built from stacked G-tetrads (G·G·G·G), which are planar cyclical assemblies of four guanine bases interacting through Hoogsteen hydrogen bonds. A G-quadruplex containing a single guanine analog substitution, such as 8-oxoguanine (O) or xanthine (X), would suffer from a loss of a Hoogsteen hydrogen bond within a G-tetrad and/or potential steric hindrance. We show that a proper arrangement of O and X bases can reestablish the hydrogen-bond pattern within a G·G·X·O tetrad. Rational incorporation of G·G·X·O tetrads in a (3+1) G-quadruplex demonstrated a similar folding topology and thermal stability to that of the unmodified G-quadruplex. pH titration conducted on X·O-modified G-quadruplexes indicated a protonation-deprotonation equilibrium of X with a pKa ∼6.7. The solution structure of a G-quadruplex containing a G·G·X·O tetrad was determined, displaying the same folding topology in both the protonated and deprotonated states. A G-quadruplex containing a deprotonated X·O pair was shown to exhibit a more electronegative groove compared to that of the unmodified one. These differences are likely to manifest in the electronic properties of G-quadruplexes and may have important implications for drug targeting and DNA-protein interactions.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-Lw-Ln), hybrid-1(3+1), UUDU
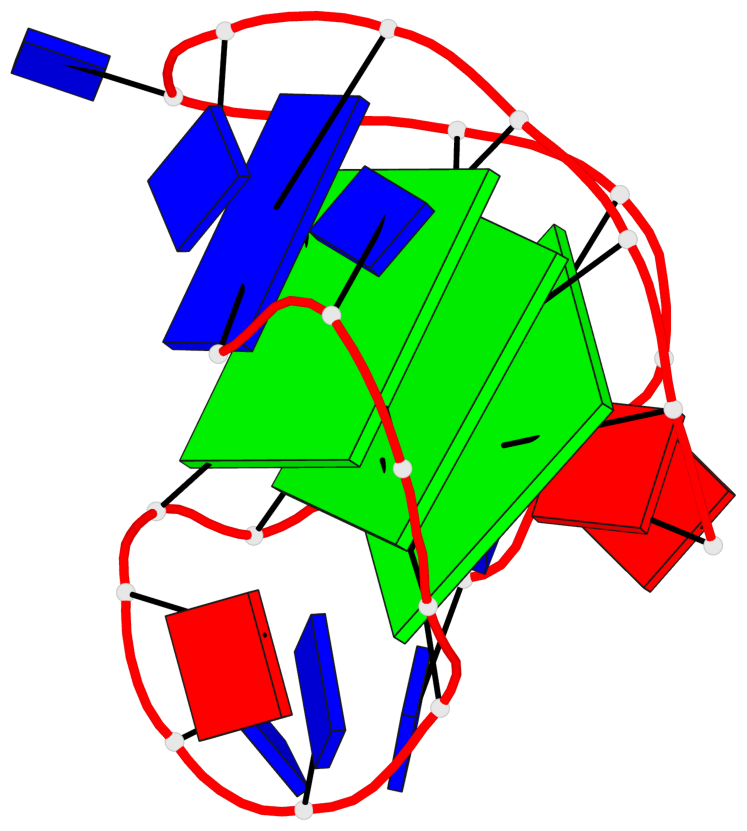
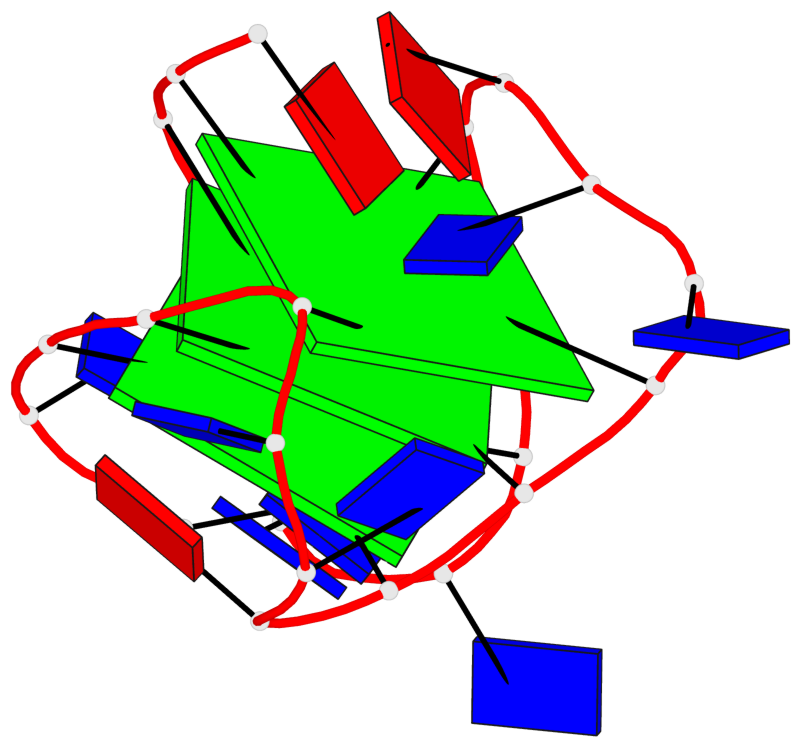
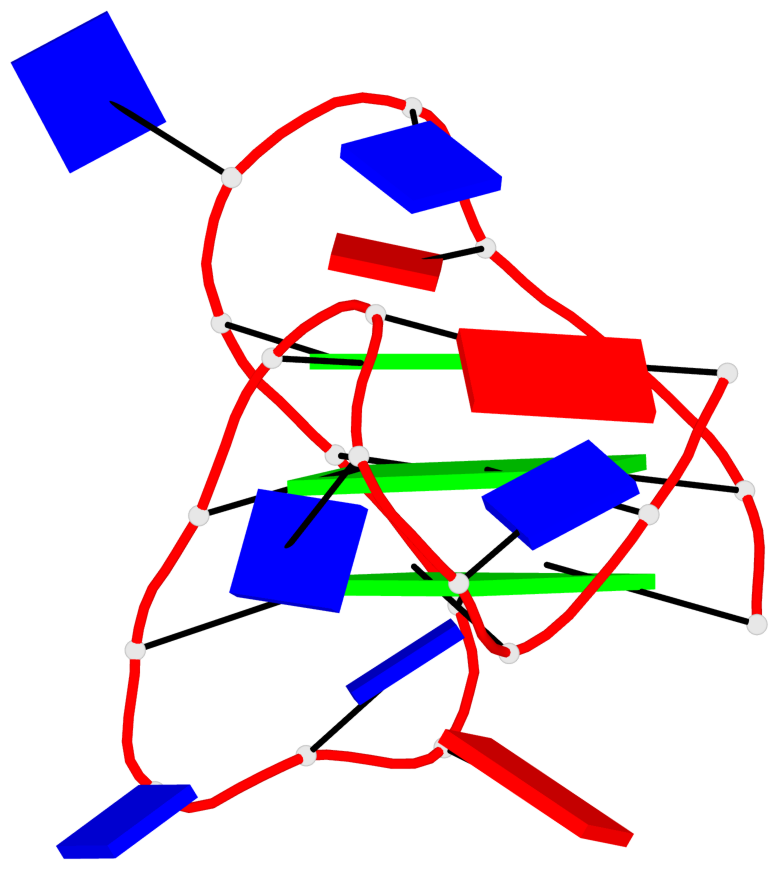
Base-block schematics in six views
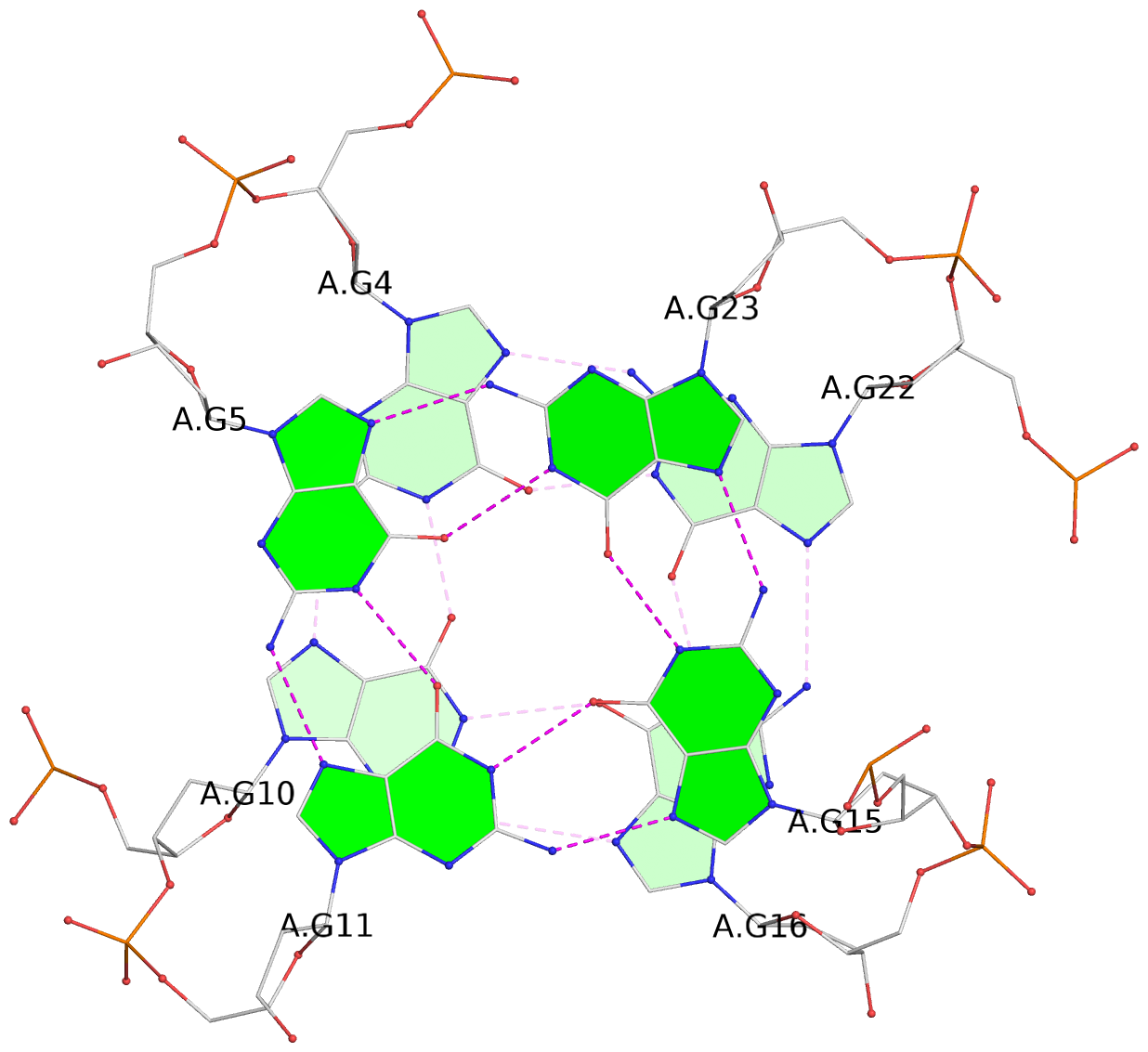
List of 3 G-tetrads
1 glyco-bond=ss-s sugar=---- groove=-wn- planarity=0.196 type=other nts=4 GggG A.DG3,A.8OG9,A.3ZO17,A.DG21 2 glyco-bond=--s- sugar=--.- groove=-wn- planarity=0.195 type=other nts=4 GGGG A.DG4,A.DG10,A.DG16,A.DG22 3 glyco-bond=--s- sugar=.--. groove=-wn- planarity=0.389 type=bowl nts=4 GGGG A.DG5,A.DG11,A.DG15,A.DG23
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.