Detailed DSSR results for the G-quadruplex: PDB entry 2n4y
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2n4y
- Class
- DNA
- Method
- NMR
- Summary
- Structure and possible function of a g-quadruplex in the long terminal repeat of the proviral hiv-1 genome
- Reference
- De Nicola B, Lech CJ, Heddi B, Regmi S, Frasson I, Perrone R, Richter SN, Phan AT (2016): "Structure and possible function of a G-quadruplex in the long terminal repeat of the proviral HIV-1 genome." Nucleic Acids Res., 44, 6442-6451. doi: 10.1093/nar/gkw432.
- Abstract
- The long terminal repeat (LTR) of the proviral human immunodeficiency virus (HIV)-1 genome is integral to virus transcription and host cell infection. The guanine-rich U3 region within the LTR promoter, previously shown to form G-quadruplex structures, represents an attractive target to inhibit HIV transcription and replication. In this work, we report the structure of a biologically relevant G-quadruplex within the LTR promoter region of HIV-1. The guanine-rich sequence designated LTR-IV forms a well-defined structure in physiological cationic solution. The nuclear magnetic resonance (NMR) structure of this sequence reveals a parallel-stranded G-quadruplex containing a single-nucleotide thymine bulge, which participates in a conserved stacking interaction with a neighboring single-nucleotide adenine loop. Transcription analysis in a HIV-1 replication competent cell indicates that the LTR-IV region may act as a modulator of G-quadruplex formation in the LTR promoter. Consequently, the LTR-IV G-quadruplex structure presented within this work could represent a valuable target for the design of HIV therapeutics.
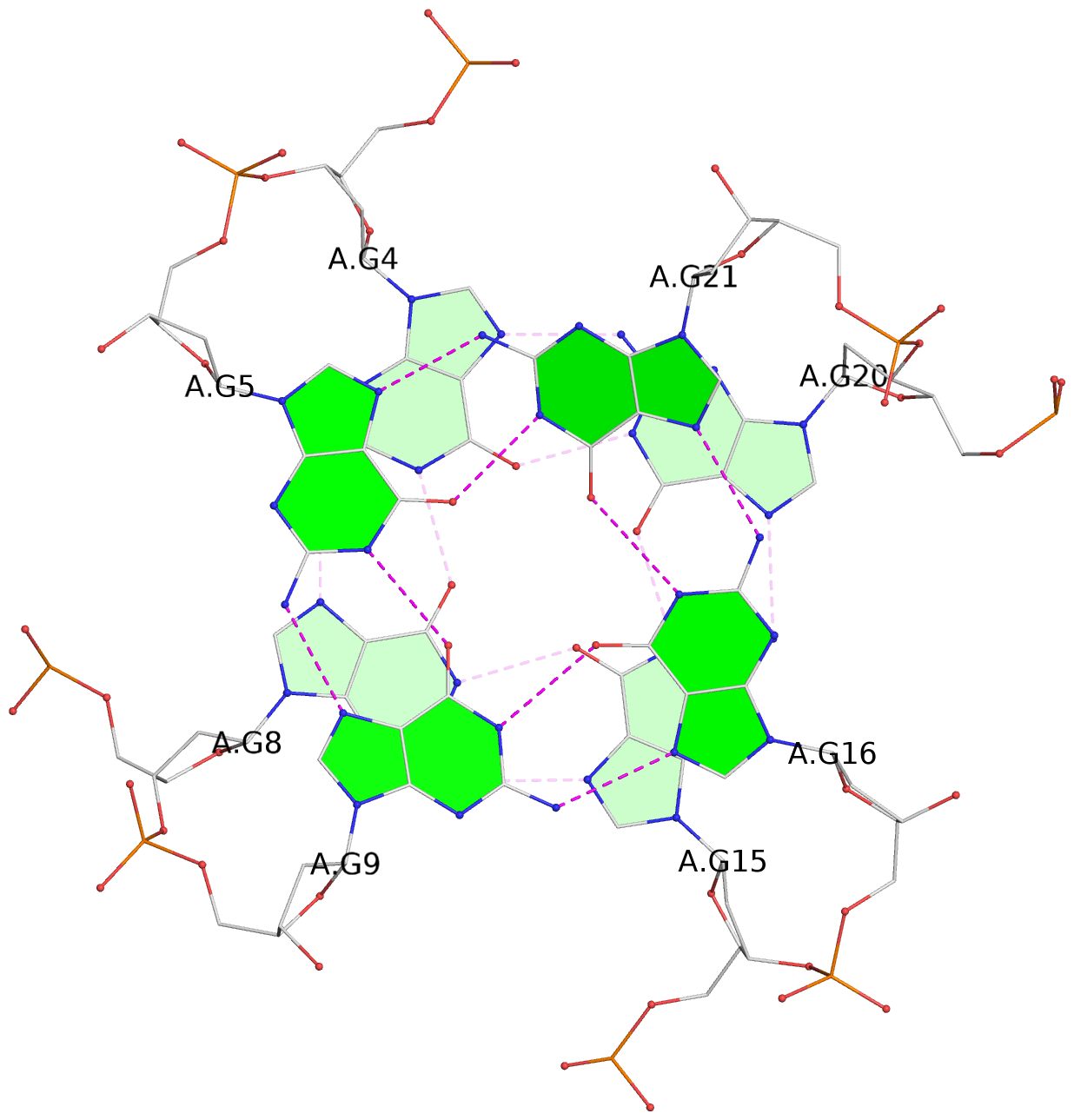
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
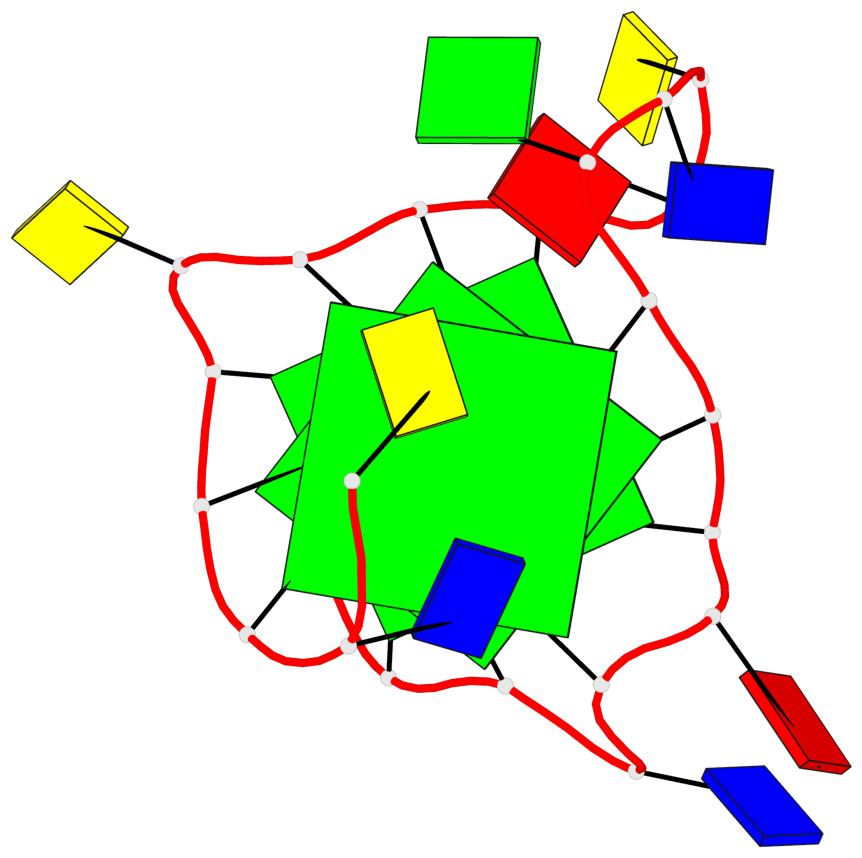
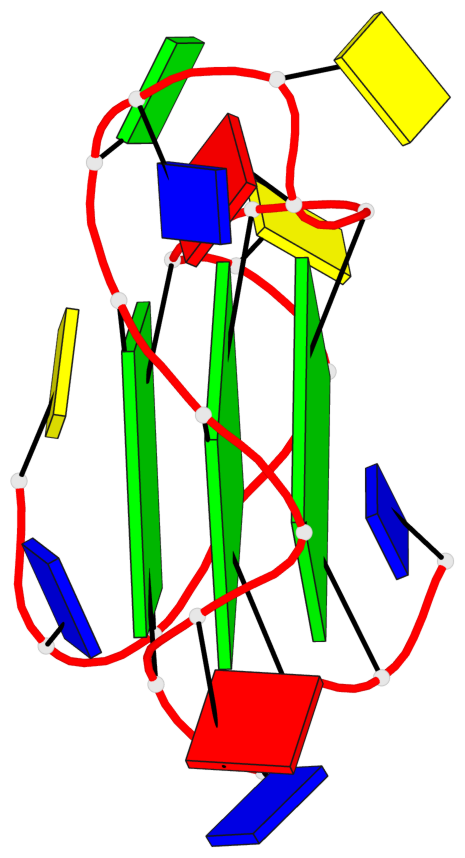
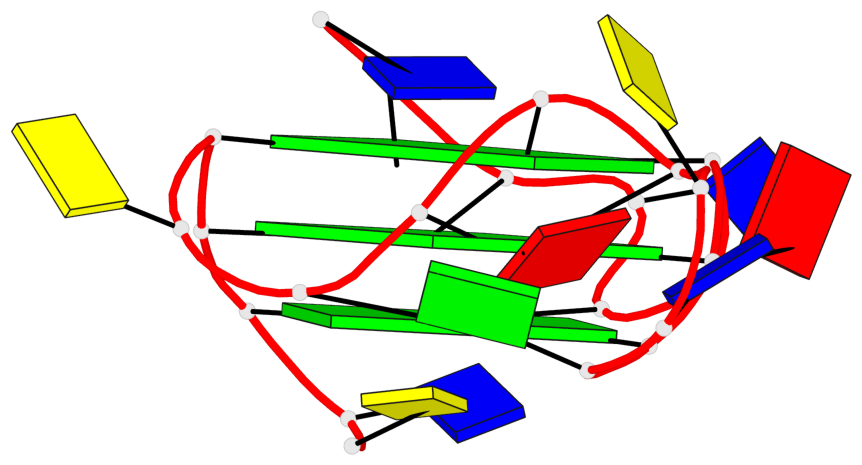
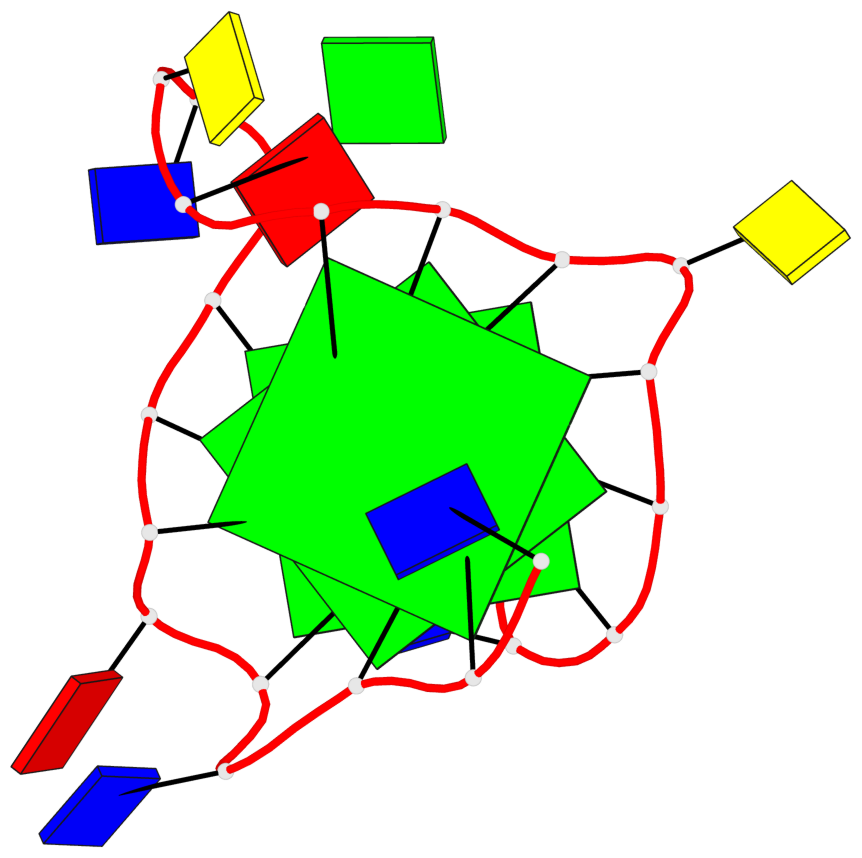
Base-block schematics in six views
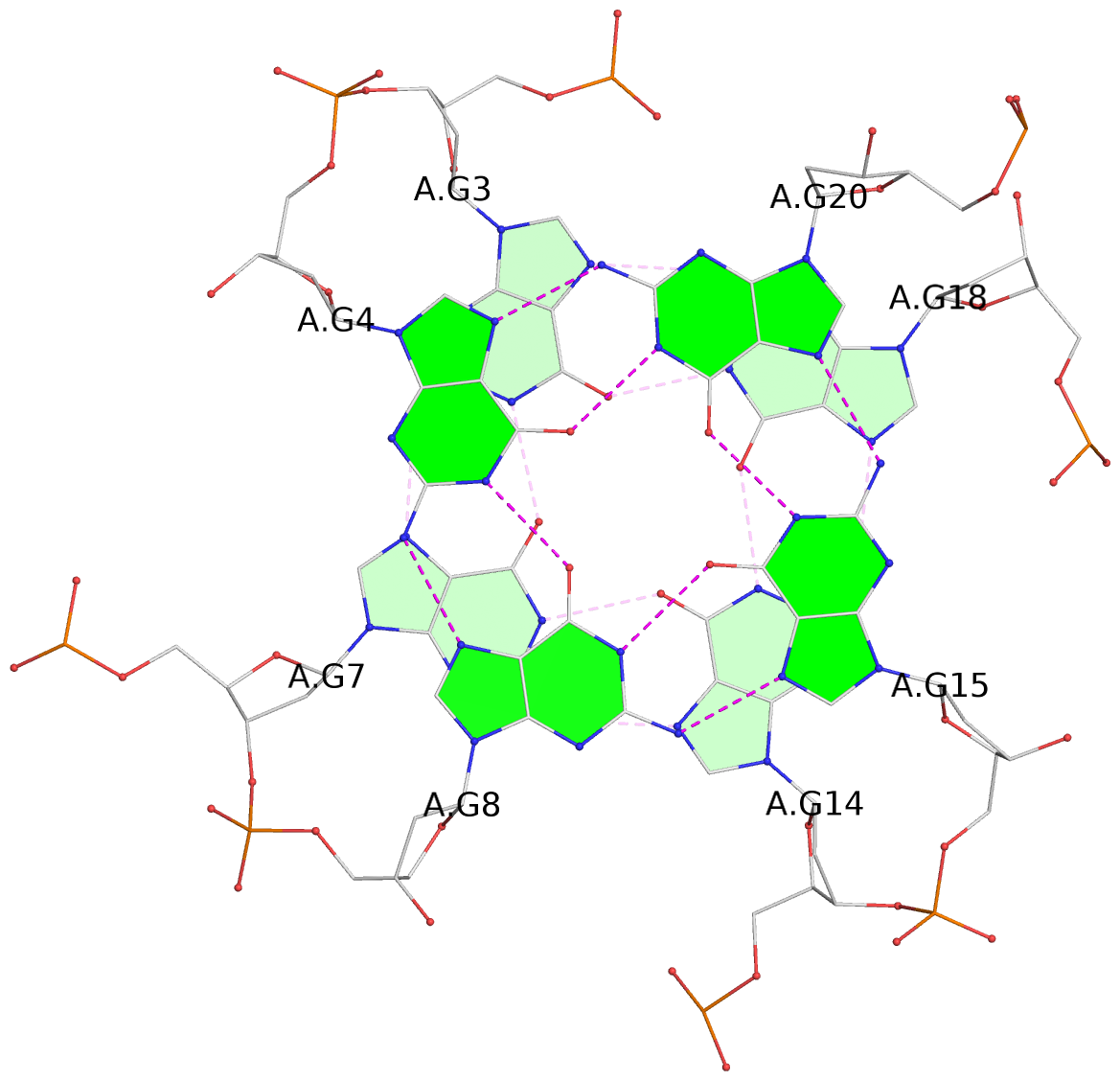
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.204 type=other nts=4 GGGG A.DG3,A.DG7,A.DG14,A.DG18 2 glyco-bond=---- sugar=---3 groove=---- planarity=0.292 type=other nts=4 GGGG A.DG4,A.DG8,A.DG15,A.DG20 3 glyco-bond=---- sugar=---- groove=---- planarity=0.479 type=bowl nts=4 GGGG A.DG5,A.DG9,A.DG16,A.DG21
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.