Detailed DSSR results for the G-quadruplex: PDB entry 2n60
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2n60
- Class
- DNA
- Method
- NMR
- Summary
- G-quadruplexes with (4n-1) guanines in the g-tetrad core: formation of a g-triad water complex and implication for small-molecule binding
- Reference
- Heddi B, Martin-Pintado N, Serimbetov Z, Kari TM, Phan AT (2016): "G-quadruplexes with (4n - 1) guanines in the G-tetrad core: formation of a G-triadwater complex and implication for small-molecule binding." Nucleic Acids Res., 44, 910-916. doi: 10.1093/nar/gkv1357.
- Abstract
- G-quadruplexes are non-canonical structures of nucleic acids, in which guanine bases form planar G-tetrads (G·G·G·G) that stack on each other in the core of the structure. G-quadruplexes generally contain multiple times of four (4n) guanines in the core. Here, we study the structure of G-quadruplexes with only (4n - 1) guanines in the core. The solution structure of a DNA sequence containing 11 guanines showed the formation of a parallel G-quadruplex involving two G-tetrads and one G-triad with a vacant site. Molecular dynamics simulation established the formation of a stable G-triad·water complex, where water molecules mimic the position of the missing guanine in the vacant site. The concept of forming G-quadruplexes with missing guanines in the core broadens the current definition of G-quadruplex-forming sequences. The potential ability of such structures to bind different metabolites, including guanine, guanosine and GTP, in the vacant site, could have biological implications in regulatory functions. Formation of this unique binding pocket in the G-triad could be used as a specific target in drug design.
- G4 notes
- 2 G-tetrads, 1 G4 helix, 1 G4 stem, 2(-P-P-P), parallel(4+0), UUUU
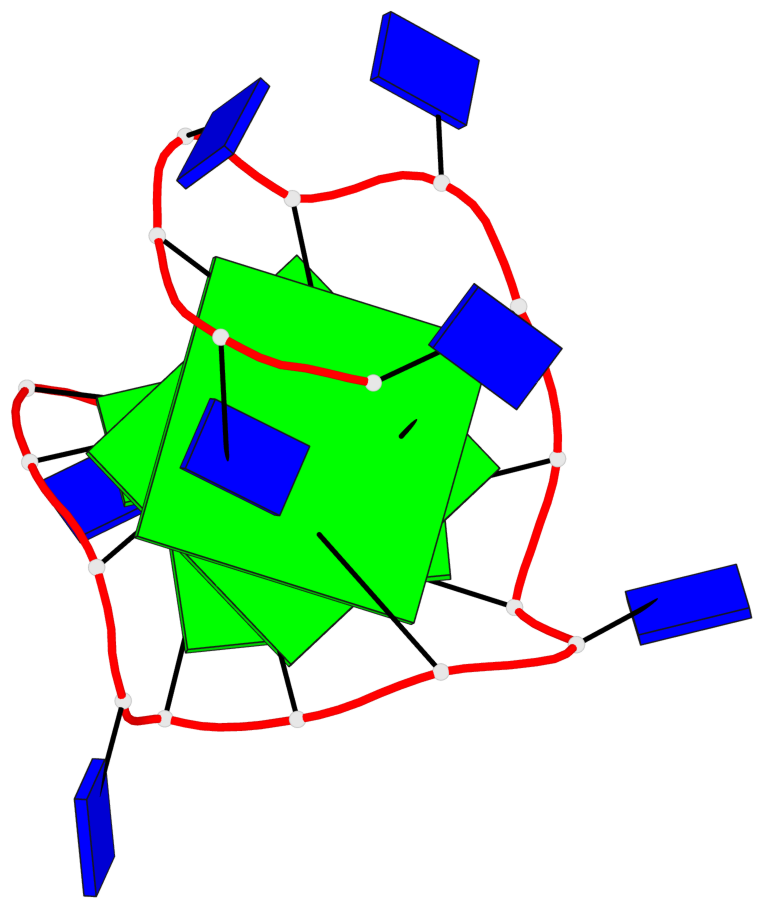
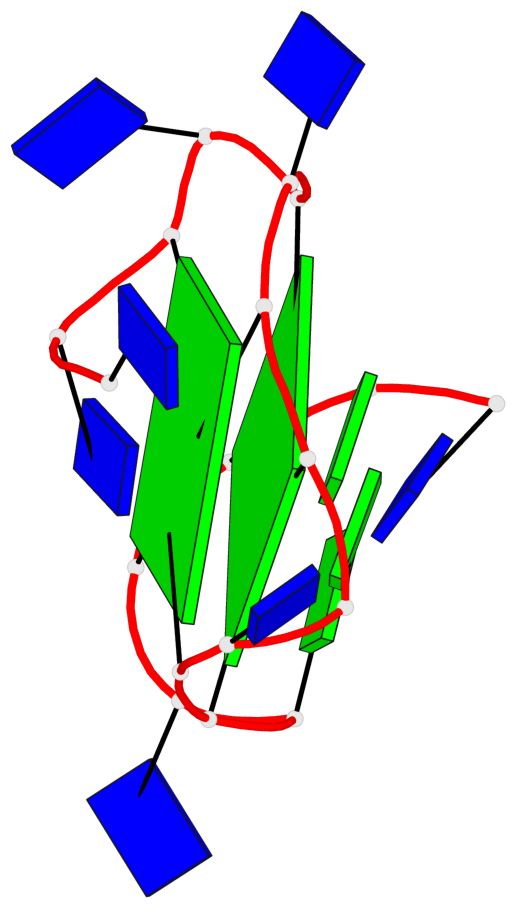
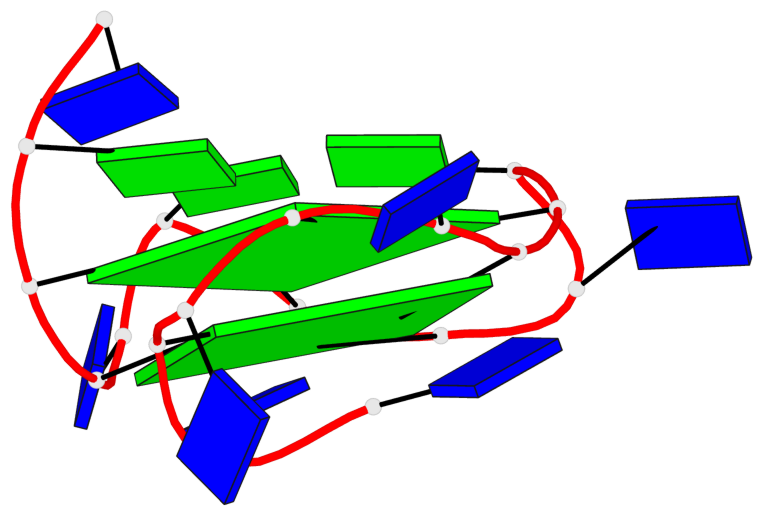
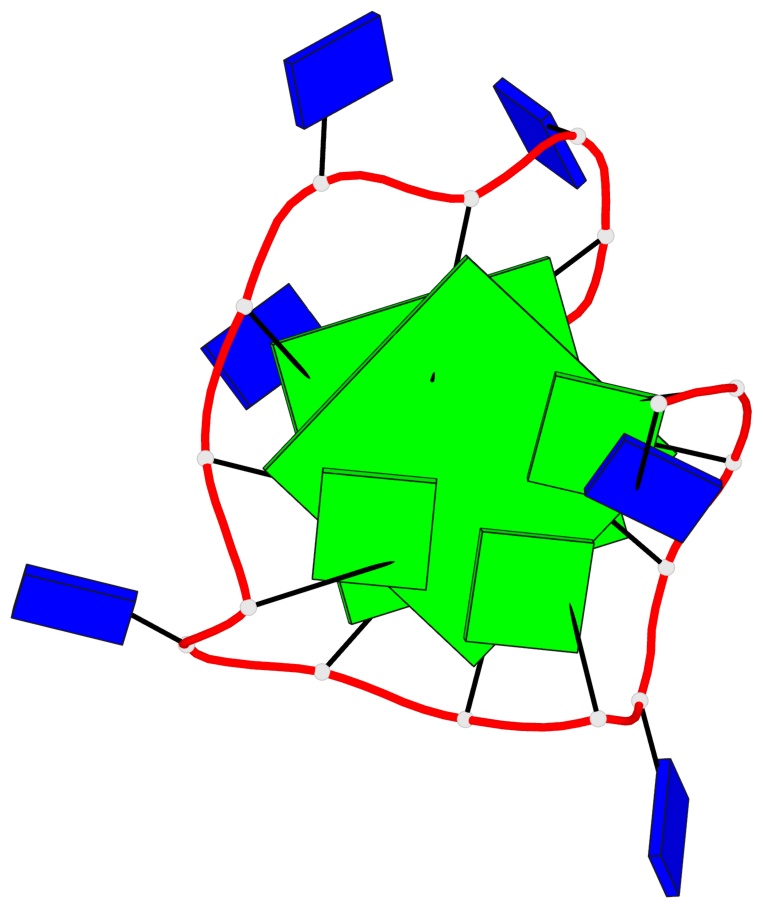
Base-block schematics in six views
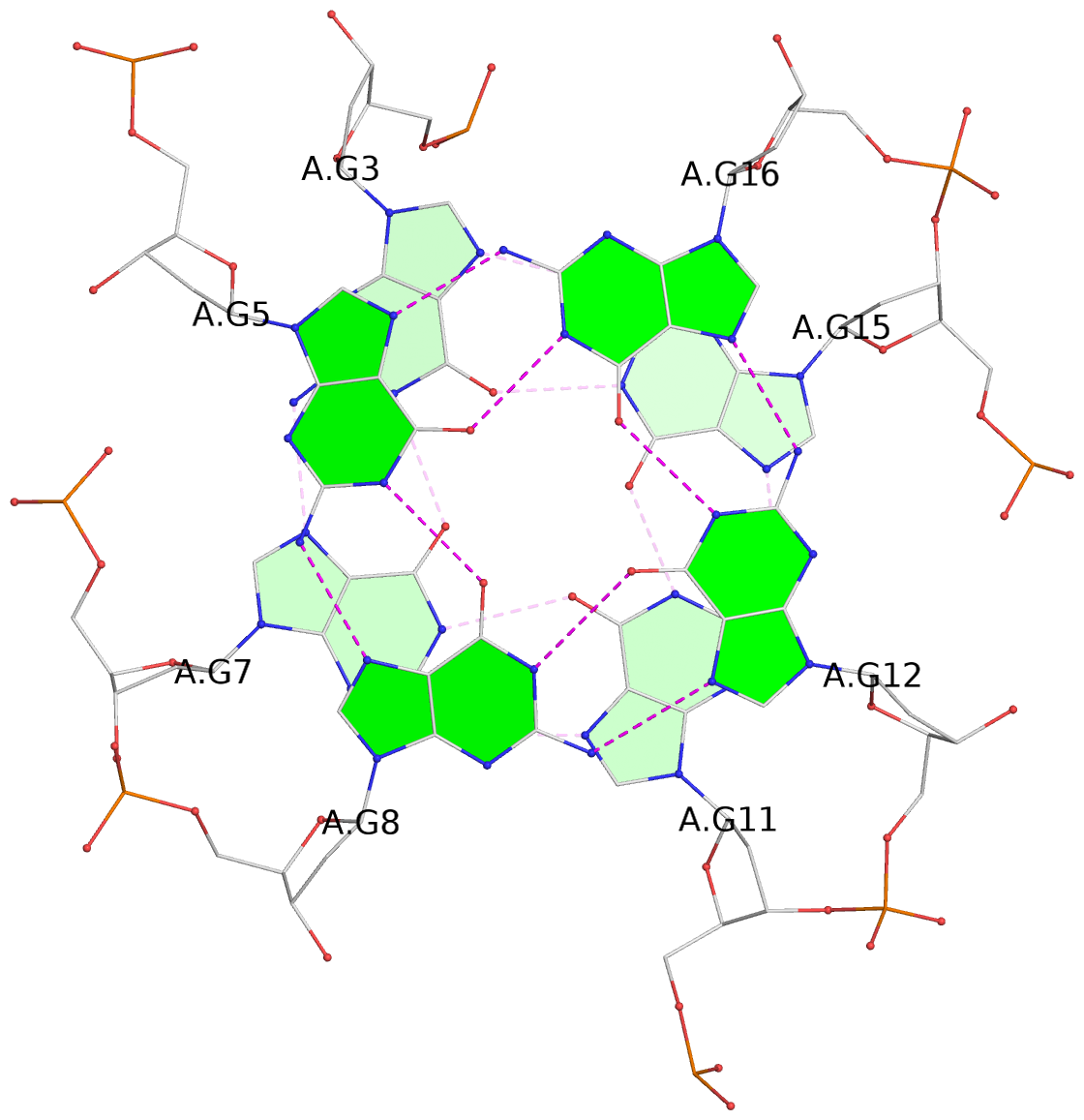
List of 2 G-tetrads
1 glyco-bond=---- sugar=.--- groove=---- planarity=0.362 type=other nts=4 GGGG A.DG3,A.DG7,A.DG11,A.DG15 2 glyco-bond=---- sugar=---- groove=---- planarity=0.362 type=other nts=4 GGGG A.DG5,A.DG8,A.DG12,A.DG16
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.