Detailed DSSR results for the G-quadruplex: PDB entry 2rqj
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2rqj
- Class
- RNA
- Method
- NMR
- Summary
- Quadruplex structure of an RNA aptamer against bovine prion protein
- Reference
- Mashima T, Matsugami A, Nishikawa F, Nishikawa S, Katahira M (2009): "Unique quadruplex structure and interaction of an RNA aptamer against bovine prion protein." Nucleic Acids Res., 37, 6249-6258. doi: 10.1093/nar/gkp647.
- Abstract
- RNA aptamers against bovine prion protein (bPrP) were obtained, most of the obtained aptamers being found to contain the r(GGAGGAGGAGGA) (R12) sequence. Then, it was revealed that R12 binds to both bPrP and its beta-isoform with high affinity. Here, we present the structure of R12. This is the first report on the structure of an RNA aptamer against prion protein. R12 forms an intramolecular parallel quadruplex. The quadruplex contains G:G:G:G tetrad and G(:A):G:G(:A):G hexad planes. Two quadruplexes form a dimer through intermolecular hexad-hexad stacking. Two lysine clusters of bPrP have been identified as binding sites for R12. The electrostatic interaction between the uniquely arranged phosphate groups of R12 and the lysine clusters is suggested to be responsible for the affinity of R12 to bPrP. The stacking interaction between the G:G:G:G tetrad planes and tryptophan residues may also contribute to the affinity. One R12 dimer molecule is supposed to simultaneously bind the two lysine clusters of one bPrP molecule, resulting in even higher affinity. The atomic coordinates of R12 would be useful for the development of R12 as a therapeutic agent against prion diseases and Alzheimer's disease.
- G4 notes
- 4 G-tetrads, 1 G4 helix, 2 G4 stems, 1 G4 coaxial stack, 2(-P-P-P), parallel(4+0), UUUU, coaxial interfaces: 5'/5'
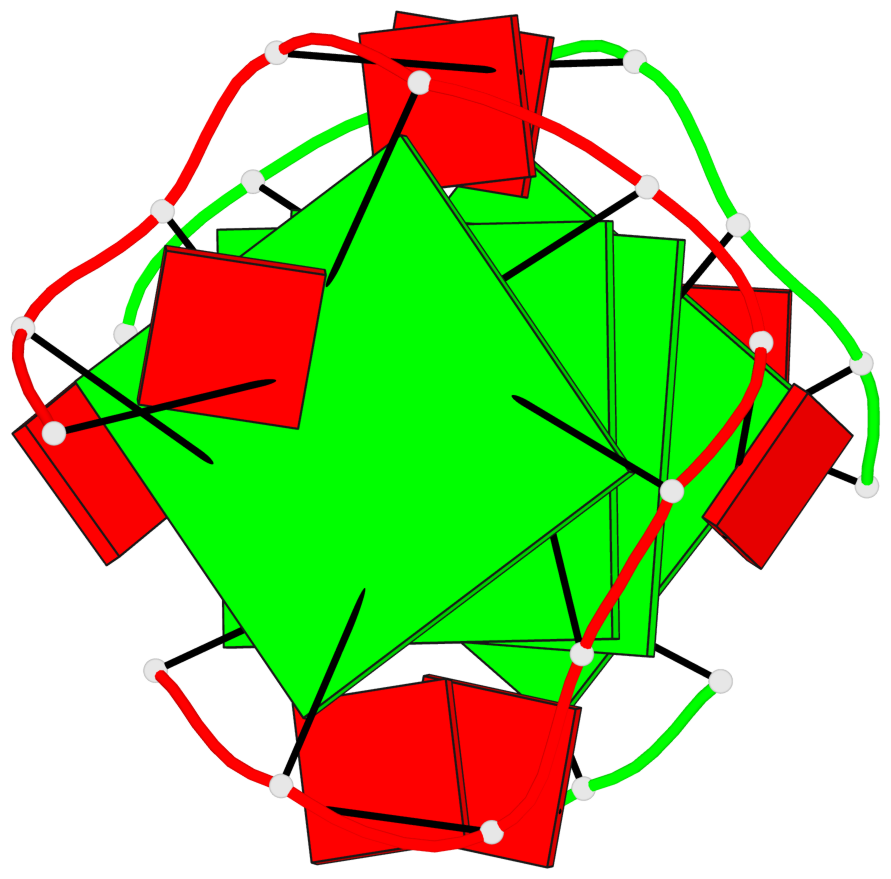
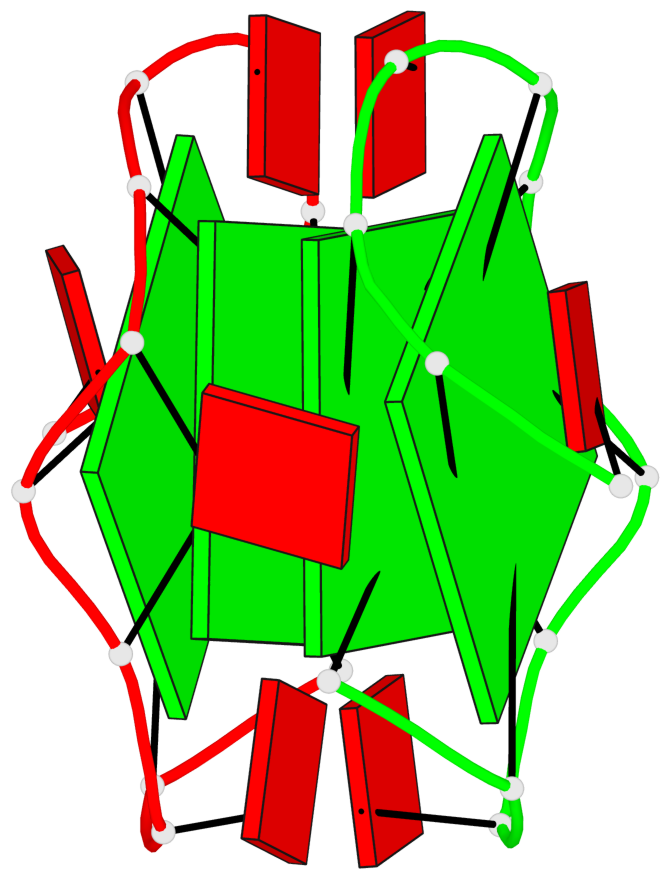
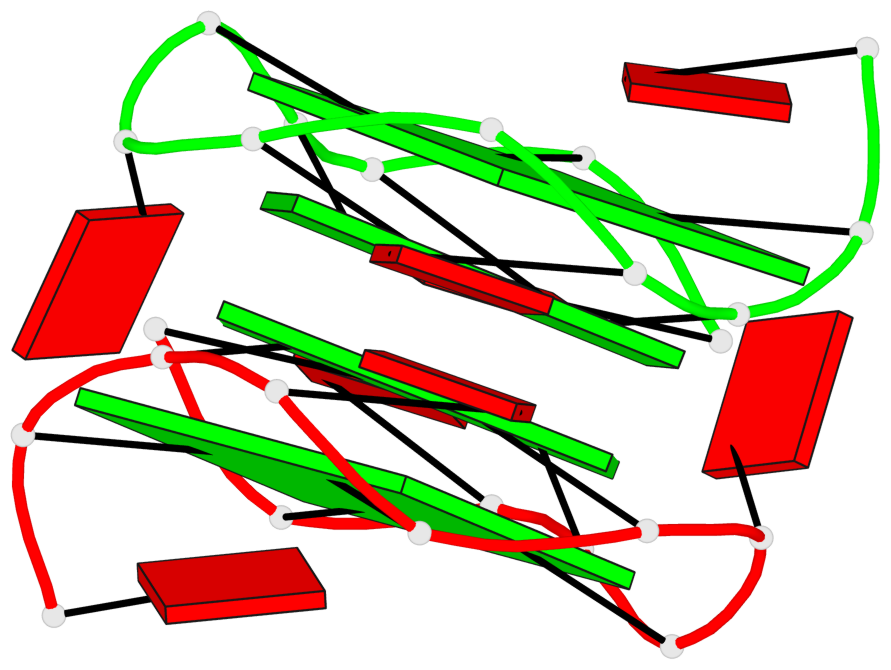
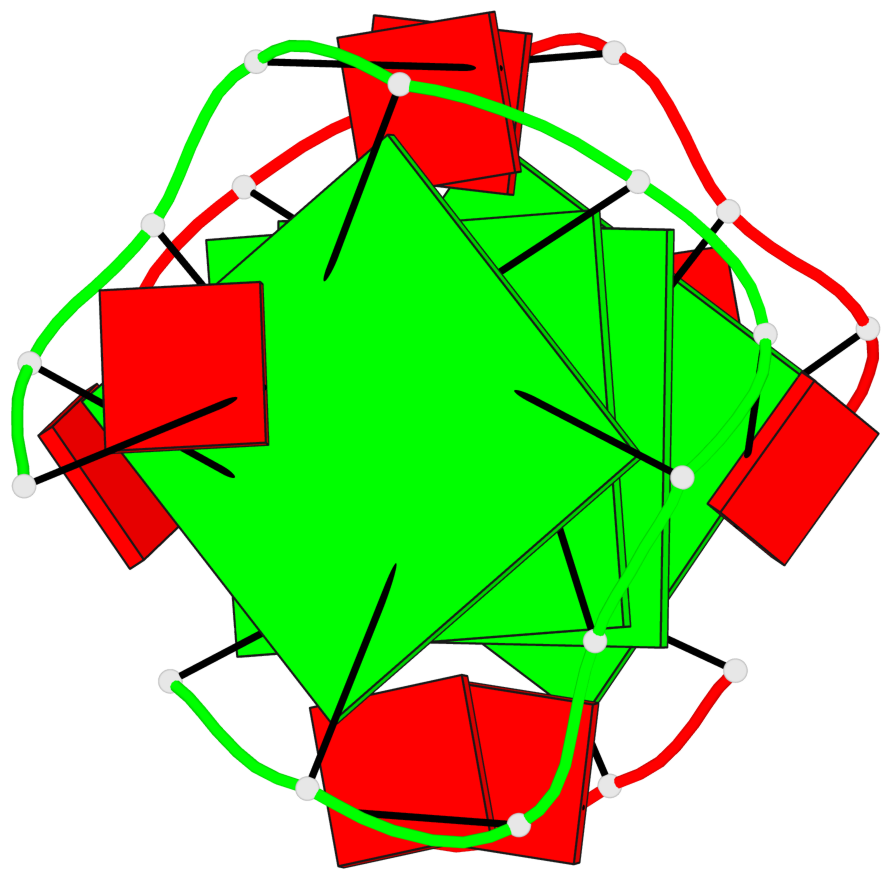
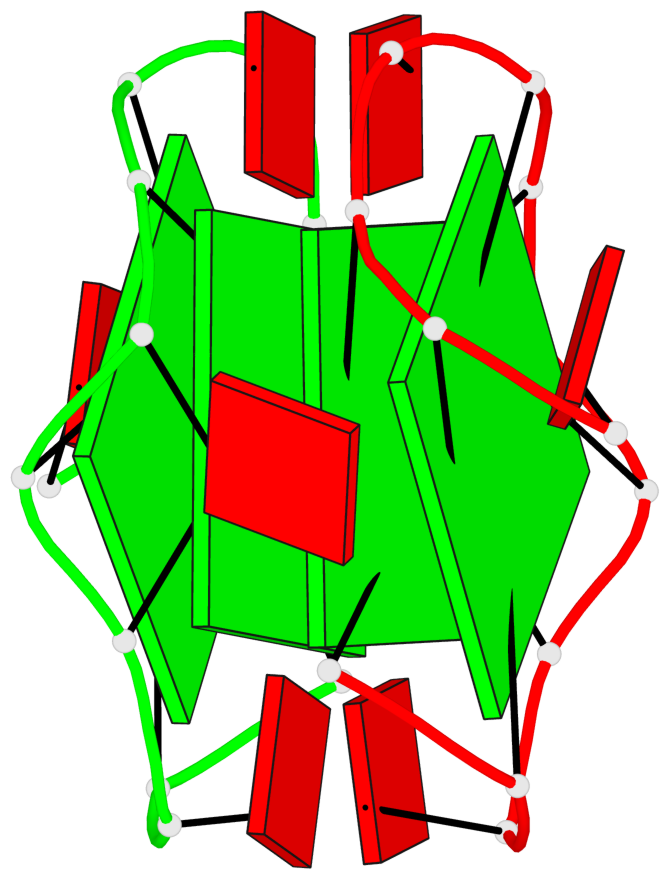
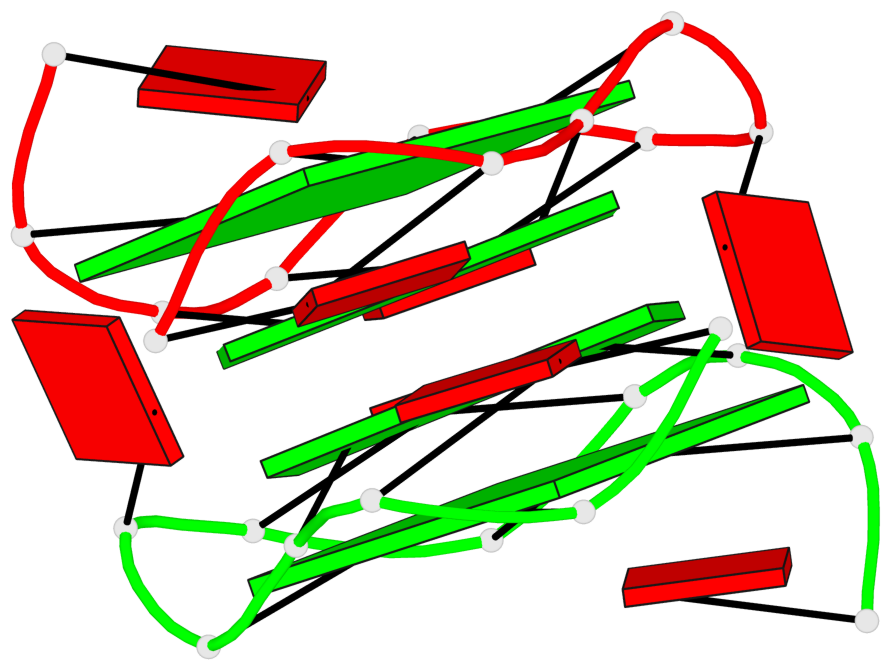
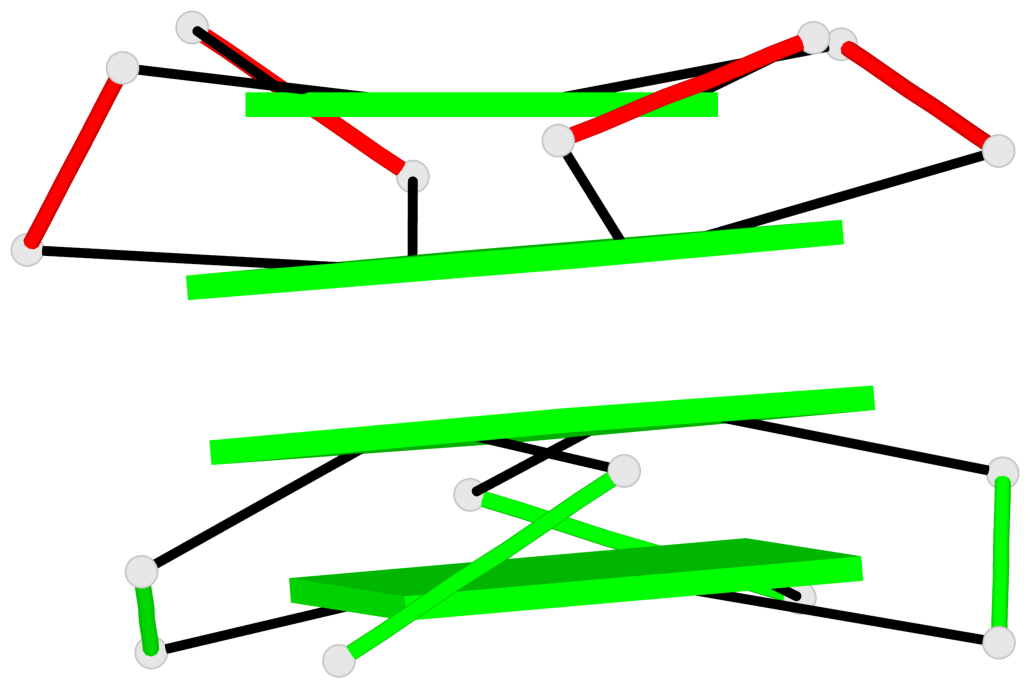
Base-block schematics in six views
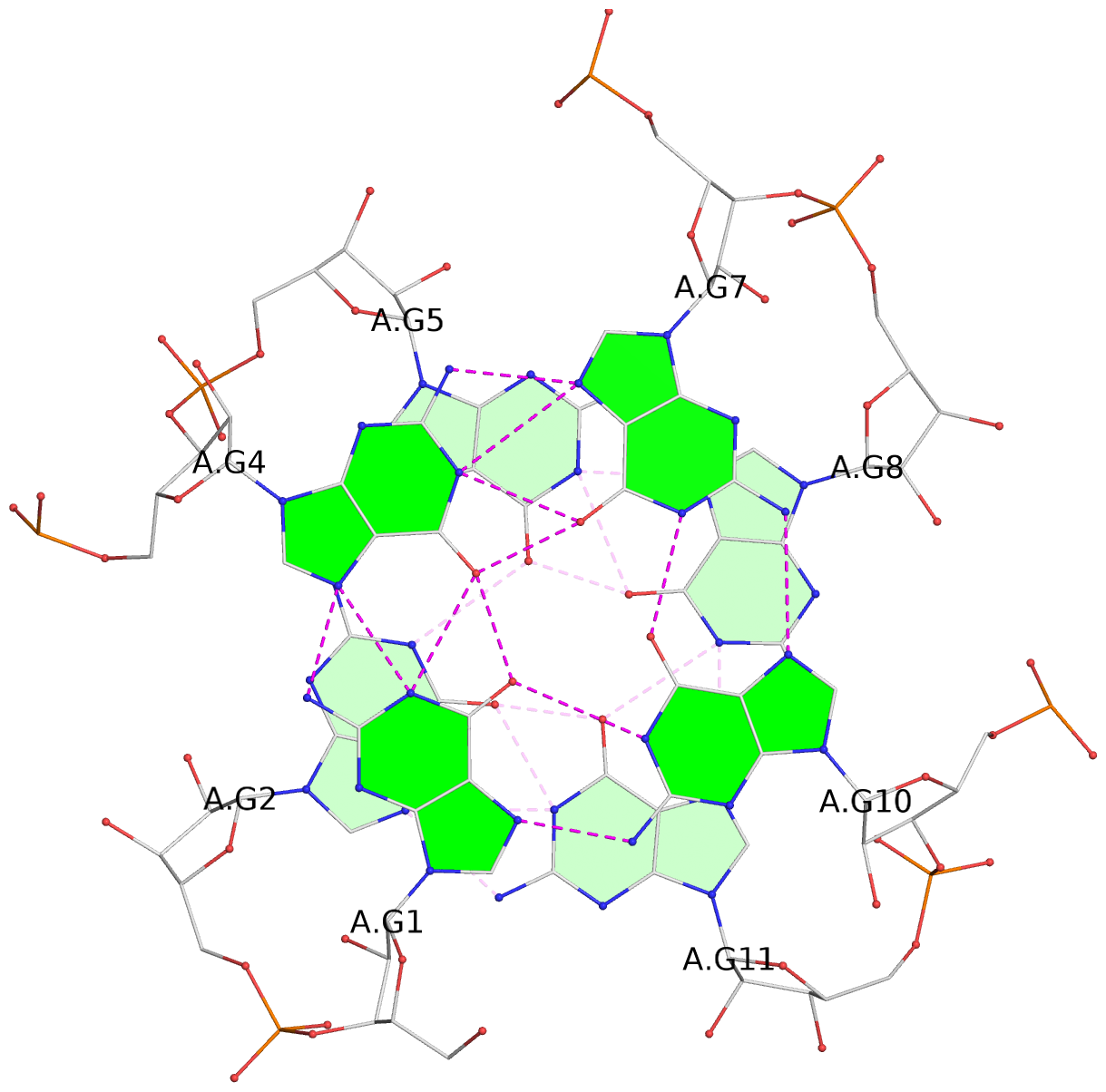
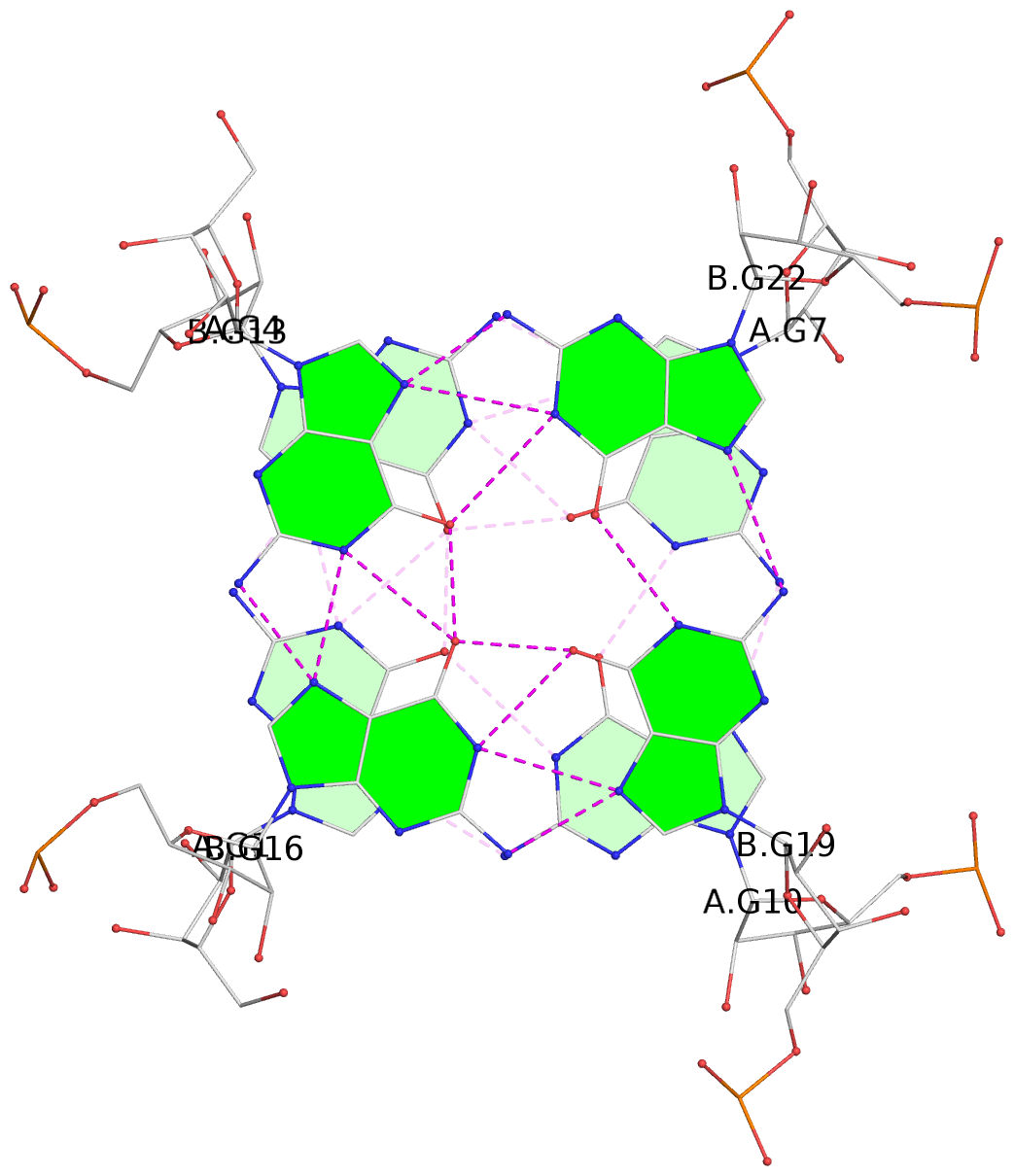
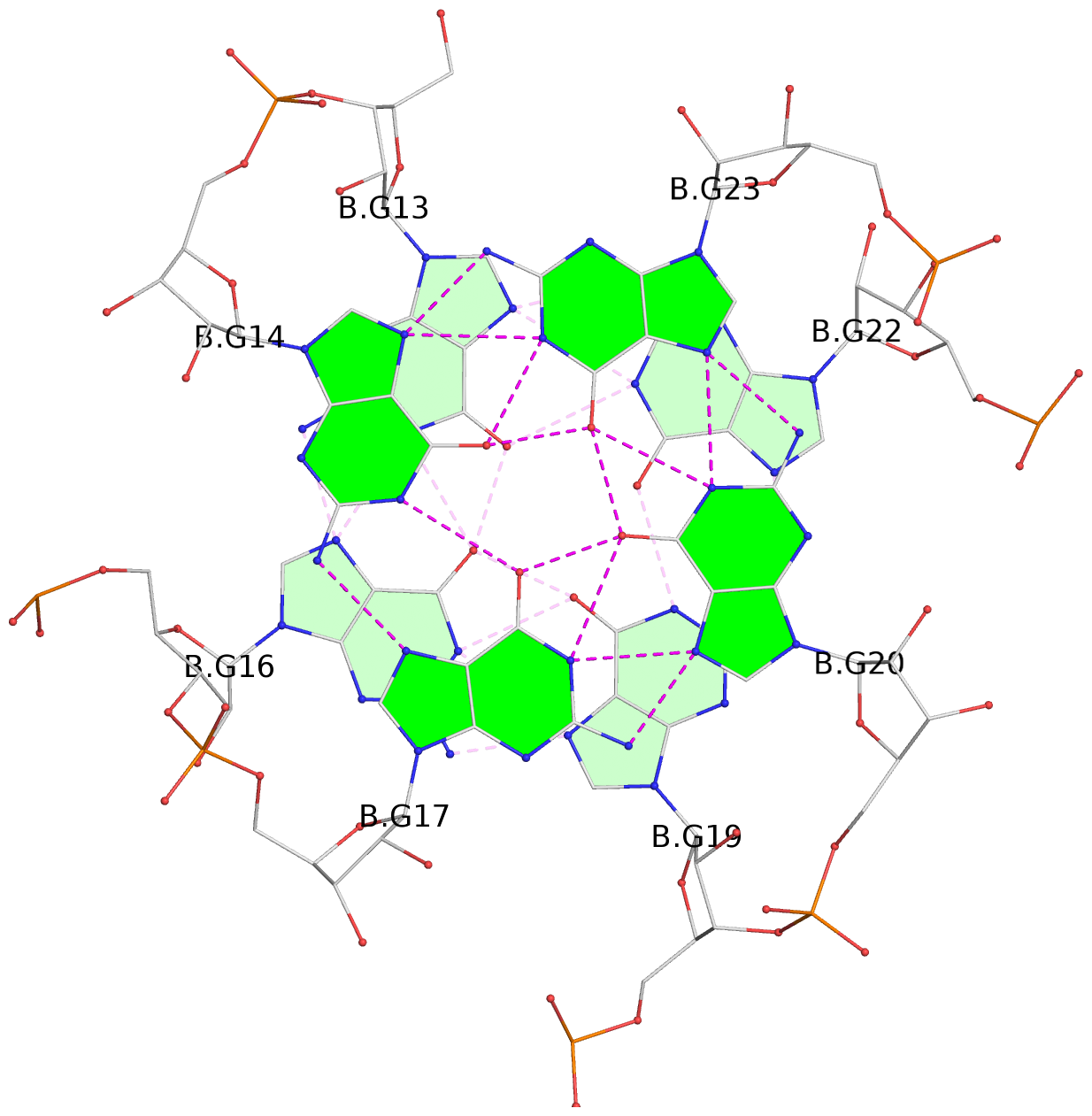
List of 4 G-tetrads
1 glyco-bond=---- sugar=-3-3 groove=---- planarity=0.092 type=planar nts=4 GGGG A.G1,A.G4,A.G7,A.G10 2 glyco-bond=---- sugar=---3 groove=---- planarity=0.076 type=planar nts=4 GGGG A.G2,A.G5,A.G8,A.G11 3 glyco-bond=---- sugar=-3-3 groove=---- planarity=0.112 type=planar nts=4 GGGG B.G13,B.G16,B.G19,B.G22 4 glyco-bond=---- sugar=---3 groove=---- planarity=0.075 type=planar nts=4 GGGG B.G14,B.G17,B.G20,B.G23
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 2 stems
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
Stem#2, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
List of 1 G4 coaxial stack
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [5'/5']