Detailed DSSR results for the G-quadruplex: PDB entry 2rsk
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2rsk
- Class
- membrane protein-RNA
- Method
- NMR
- Summary
- RNA aptamer against prion protein in complex with the partial binding peptide
- Reference
- Mashima T, Nishikawa F, Kamatari YO, Fujiwara H, Saimura M, Nagata T, Kodaki T, Nishikawa S, Kuwata K, Katahira M (2013): "Anti-prion activity of an RNA aptamer and its structural basis." Nucleic Acids Res., 41, 1355-1362. doi: 10.1093/nar/gks1132.
- Abstract
- Prion proteins (PrPs) cause prion diseases, such as bovine spongiform encephalopathy. The conversion of a normal cellular form (PrP(C)) of PrP into an abnormal form (PrP(Sc)) is thought to be associated with the pathogenesis. An RNA aptamer that tightly binds to and stabilizes PrP(C) is expected to block this conversion and to thereby prevent prion diseases. Here, we show that an RNA aptamer comprising only 12 residues, r(GGAGGAGGAGGA) (R12), reduces the PrP(Sc) level in mouse neuronal cells persistently infected with the transmissible spongiform encephalopathy agent. Nuclear magnetic resonance analysis revealed that R12, folded into a unique quadruplex structure, forms a dimer and that each monomer simultaneously binds to two portions of the N-terminal half of PrP(C), resulting in tight binding. Electrostatic and stacking interactions contribute to the affinity of each portion. Our results demonstrate the therapeutic potential of an RNA aptamer as to prion diseases.
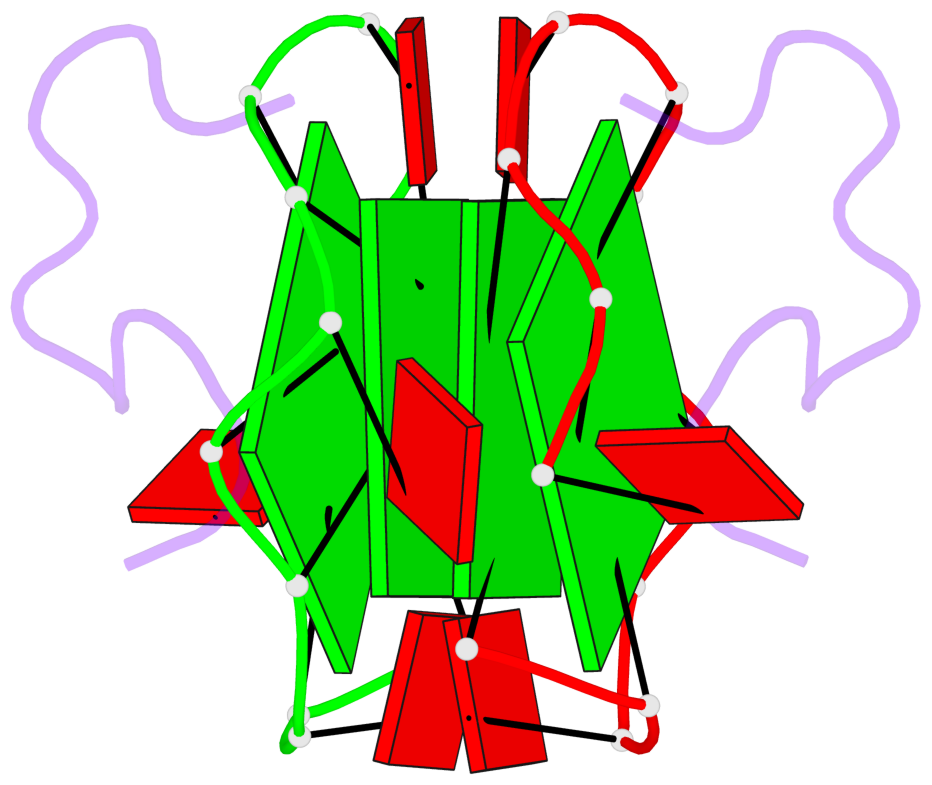
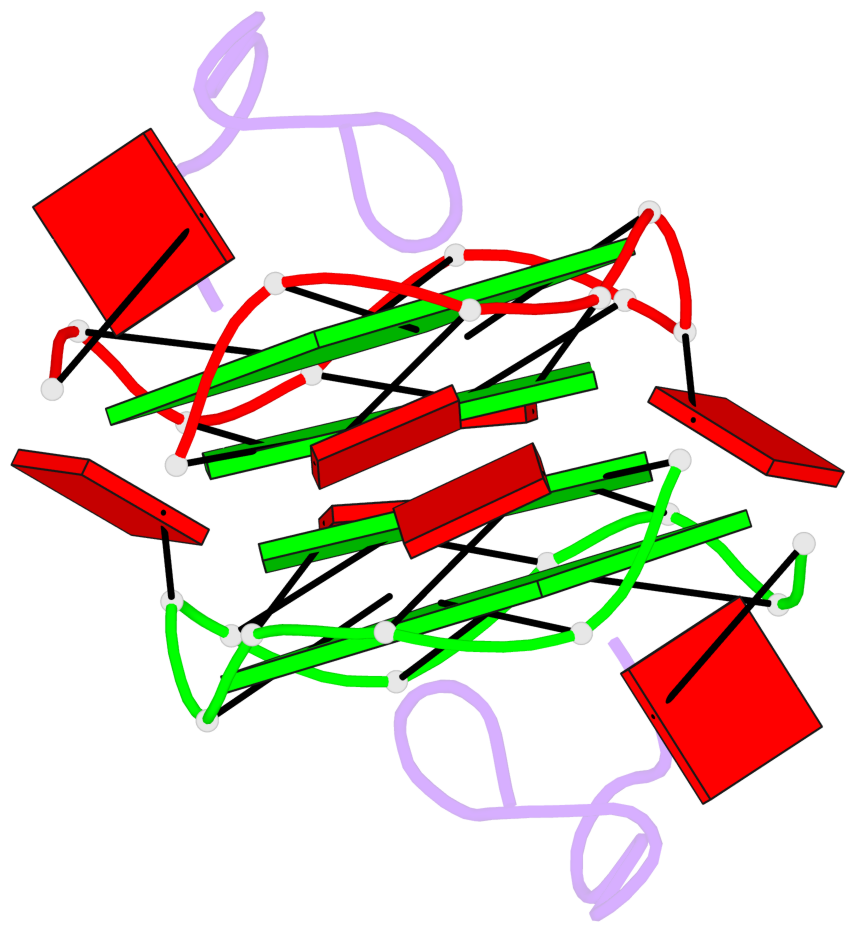
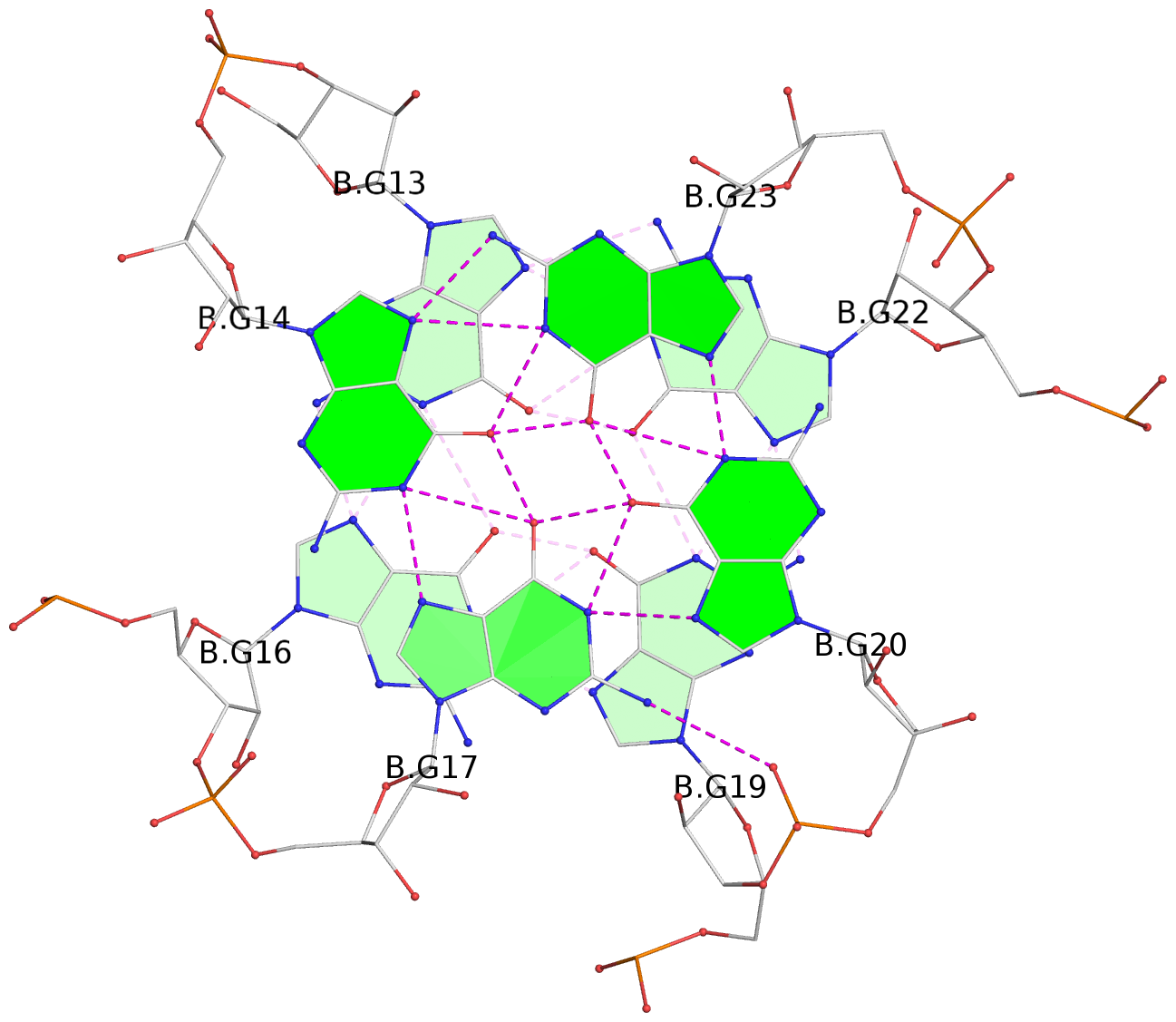
- G4 notes
- 4 G-tetrads, 1 G4 helix, 2 G4 stems, 1 G4 coaxial stack, 2(-P-P-P), parallel(4+0), UUUU, coaxial interfaces: 5'/5'
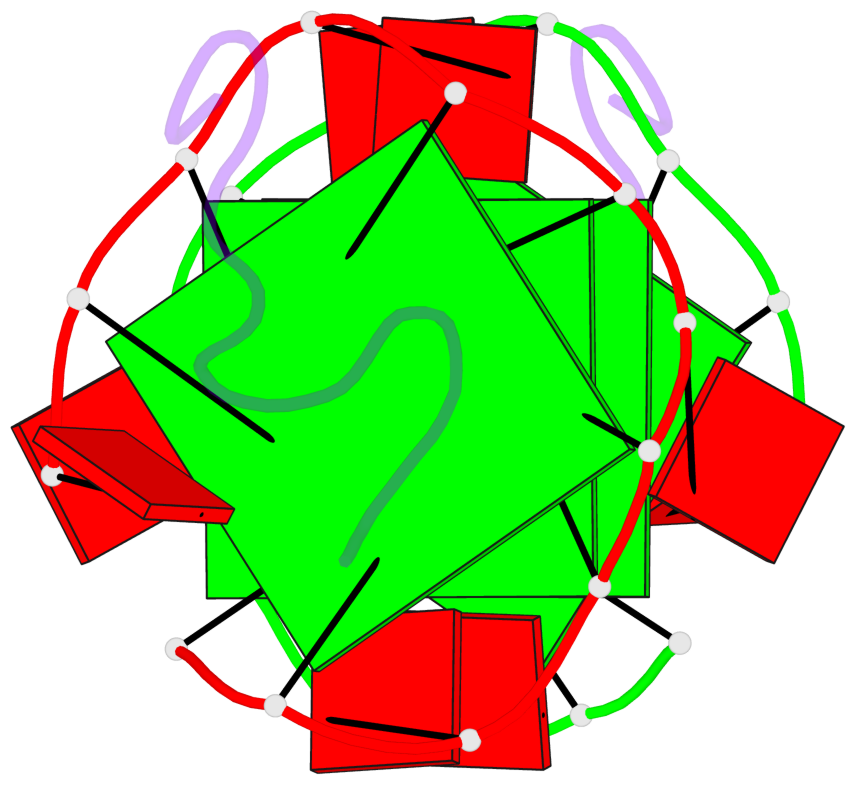
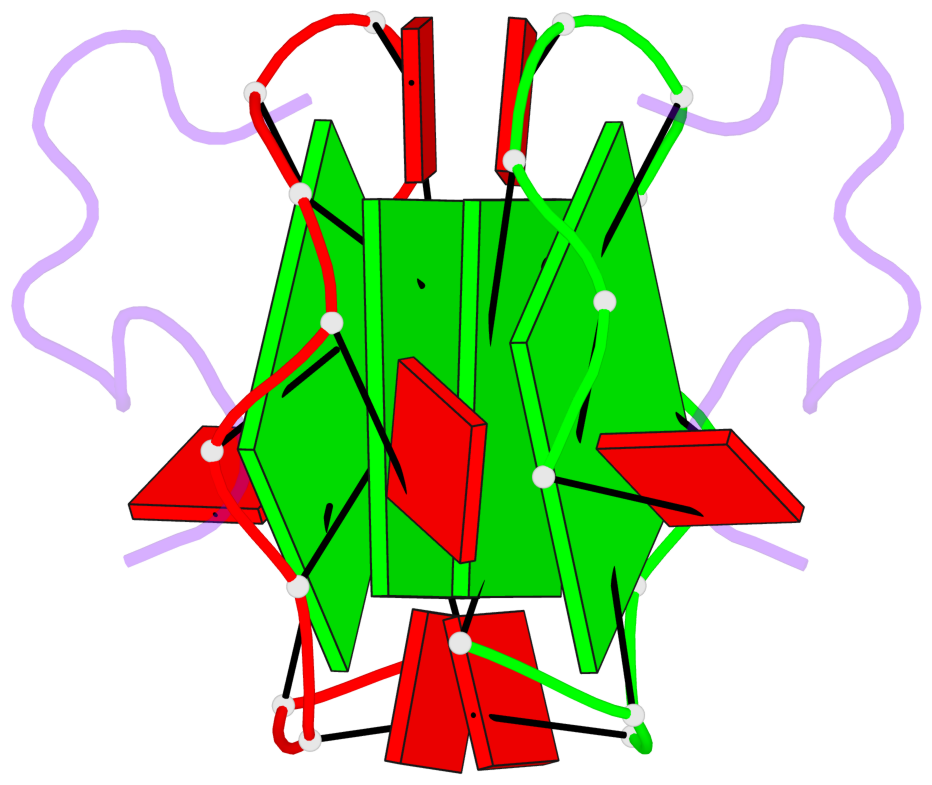
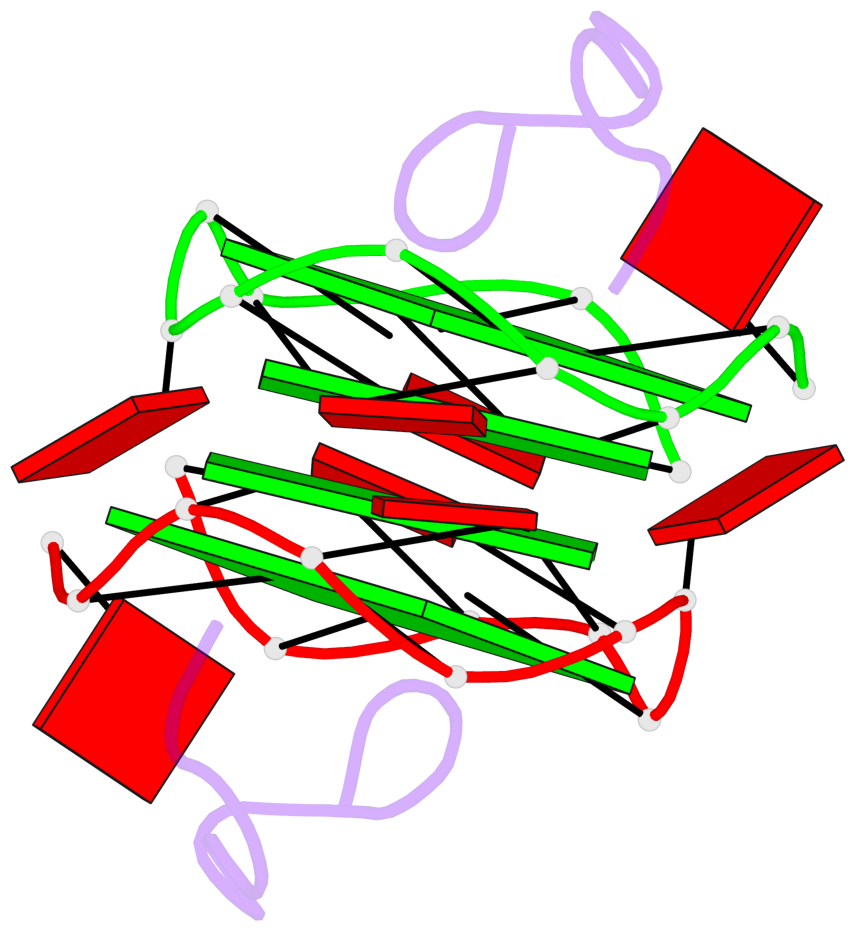
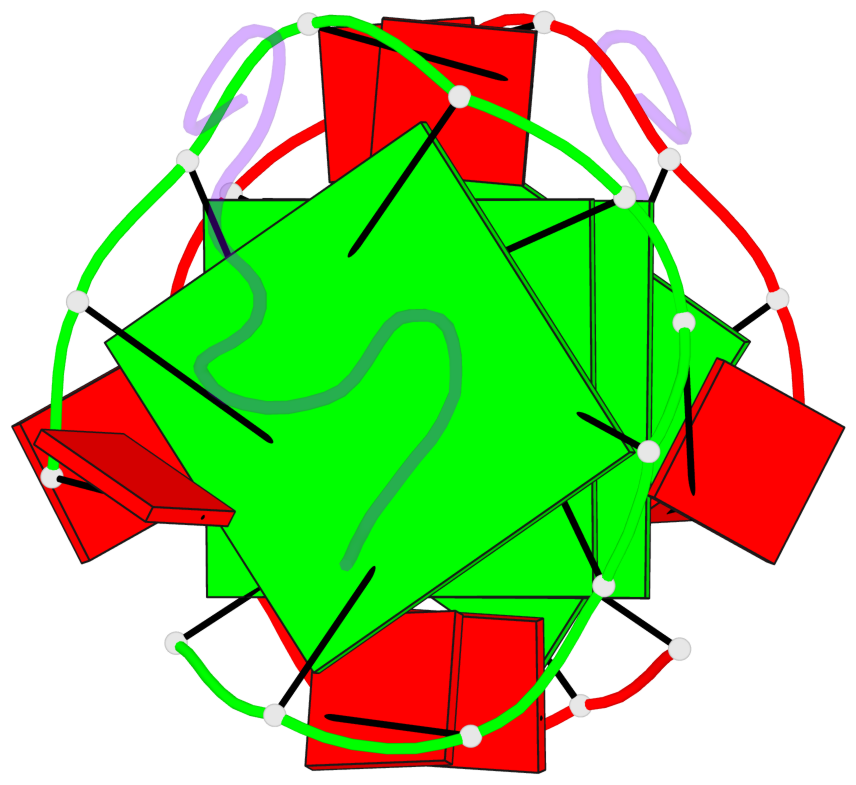
Base-block schematics in six views
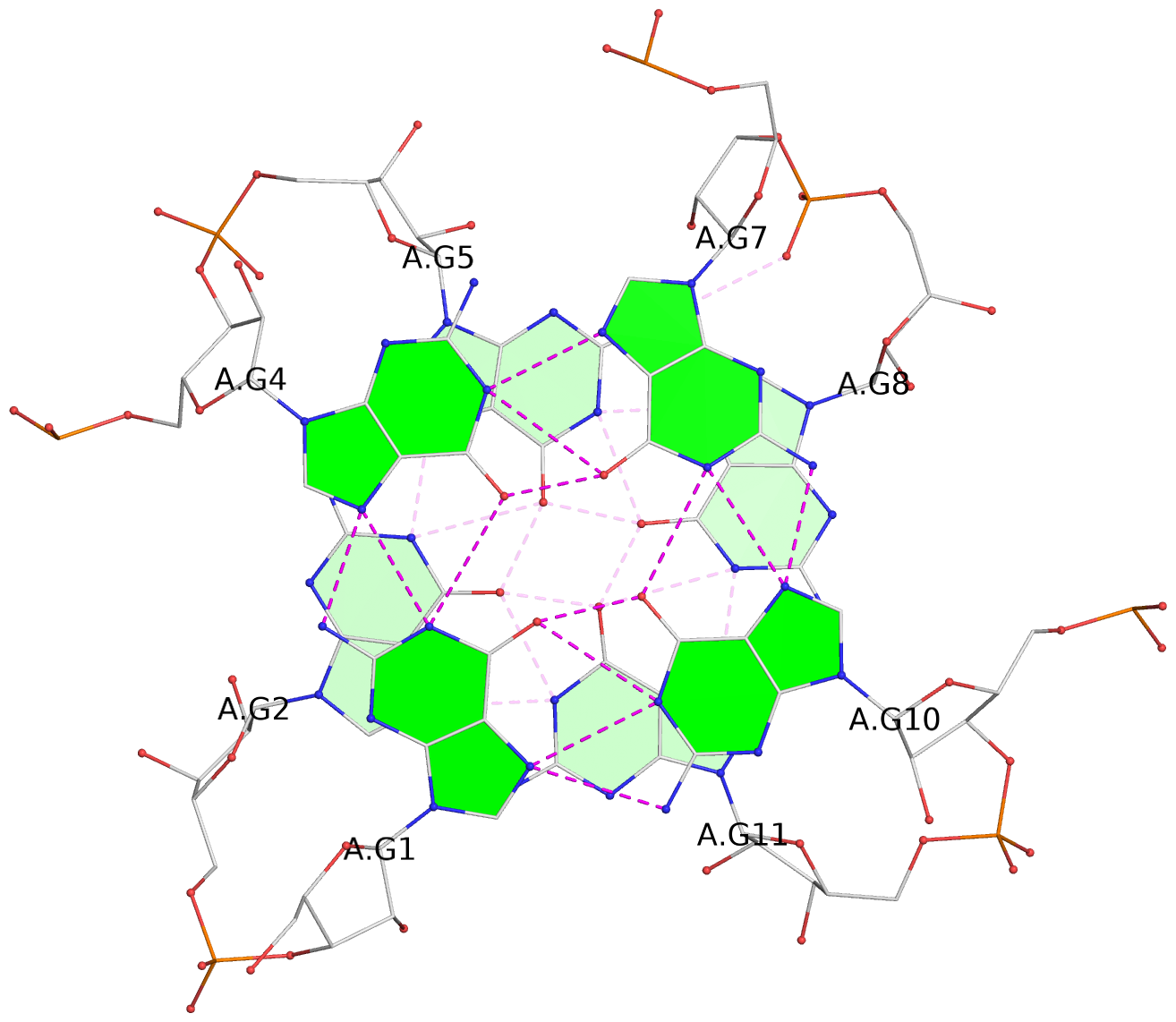
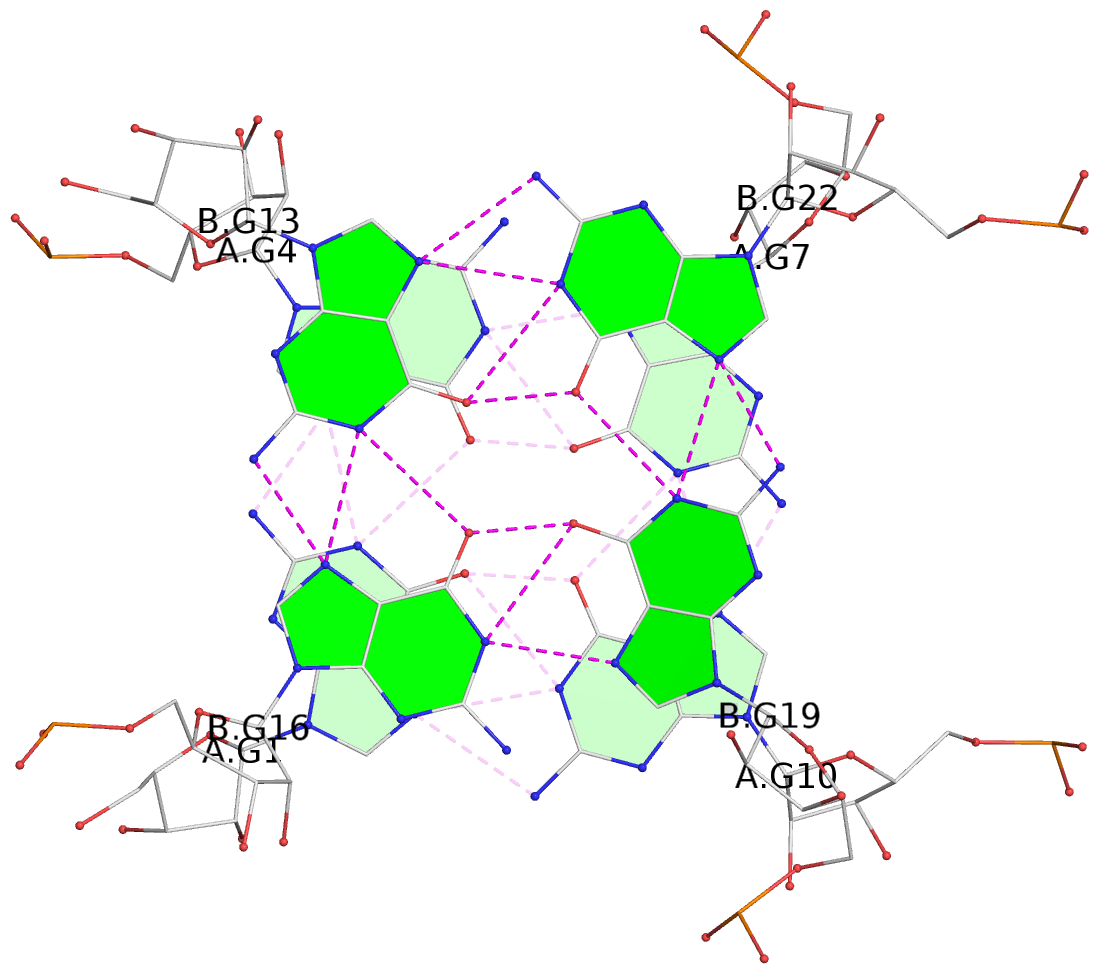
List of 4 G-tetrads
1 glyco-bond=---- sugar=-3-3 groove=---- planarity=0.264 type=other nts=4 GGGG A.G1,A.G4,A.G7,A.G10 2 glyco-bond=---- sugar=---. groove=---- planarity=0.371 type=bowl nts=4 GGGG A.G2,A.G5,A.G8,A.G11 3 glyco-bond=---- sugar=-3-3 groove=---- planarity=0.269 type=other nts=4 GGGG B.G13,B.G16,B.G19,B.G22 4 glyco-bond=---- sugar=---. groove=---- planarity=0.365 type=bowl nts=4 GGGG B.G14,B.G17,B.G20,B.G23
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 2 stems
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
Stem#2, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
List of 1 G4 coaxial stack
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [5'/5']