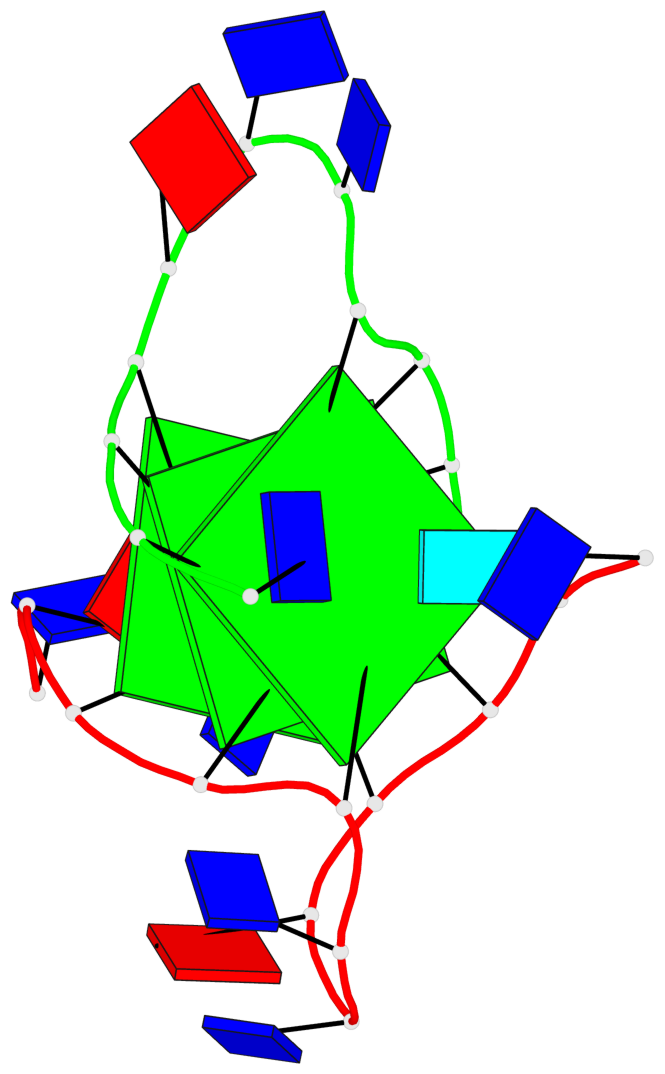
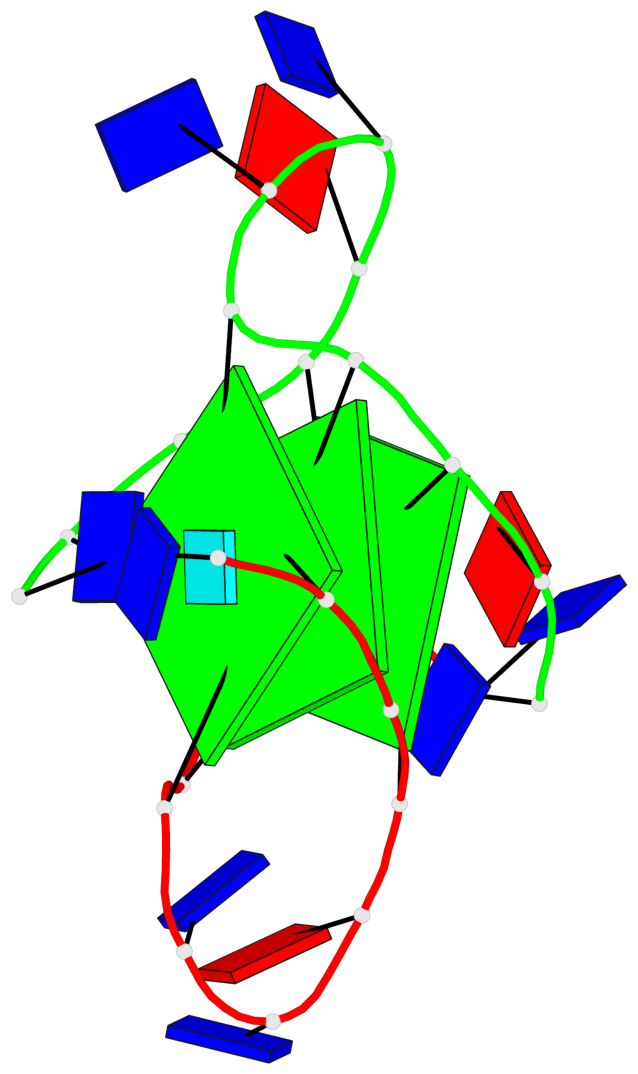
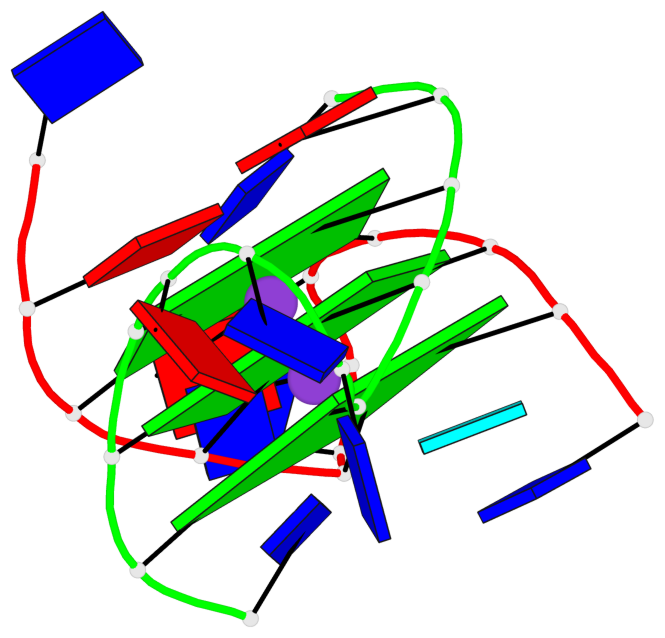
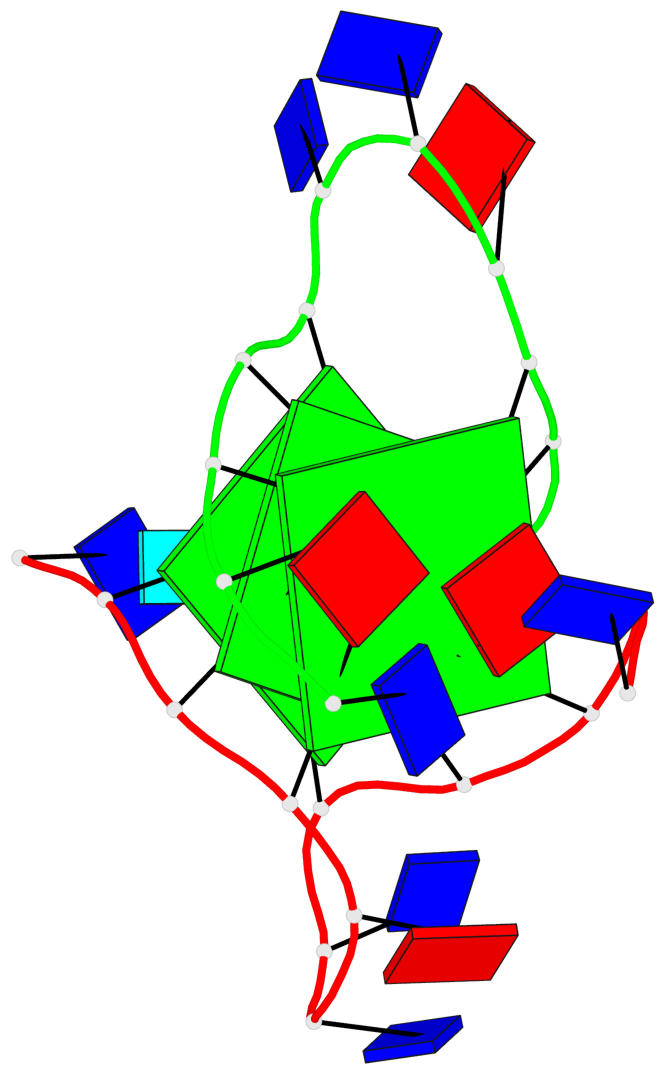
Detailed DSSR results for the G-quadruplex: PDB entry 3ce5
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 3ce5
- Class
- DNA
- Method
- X-ray (2.5 Å)
- Summary
- A bimolecular parallel-stranded human telomeric quadruplex in complex with a 3,6,9-trisubstituted acridine molecule braco19
- Reference
- Campbell NH, Parkinson GN, Reszka AP, Neidle S (2008): "Structural basis of DNA quadruplex recognition by an acridine drug." J.Am.Chem.Soc., 130, 6722-6724. doi: 10.1021/ja8016973.
- Abstract
- The crystal structure of a complex between the bimolecular human telomeric quadruplex d(TAGGGTTAGGGT)2 and the experimental anticancer drug BRACO-19, has been determined, to 2.5 A resolution. The binding site for the BRACO-19 molecule is at the interface of two parallel-folded quadruplexes, sandwiched between a G-tetrad surface and a TATA tetrad, and held in the site by networks of water molecules. The structure rationalizes the existing structure-activity data and provides a starting-point for the structure-based design of quadruplex-binding ligands
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, parallel(4+0), UUUU
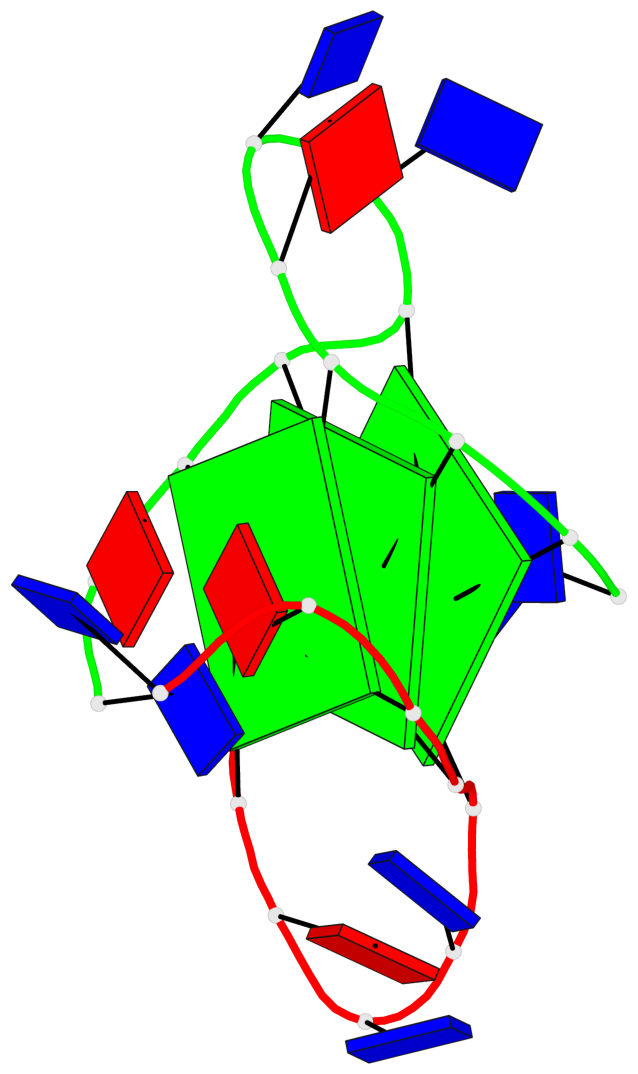
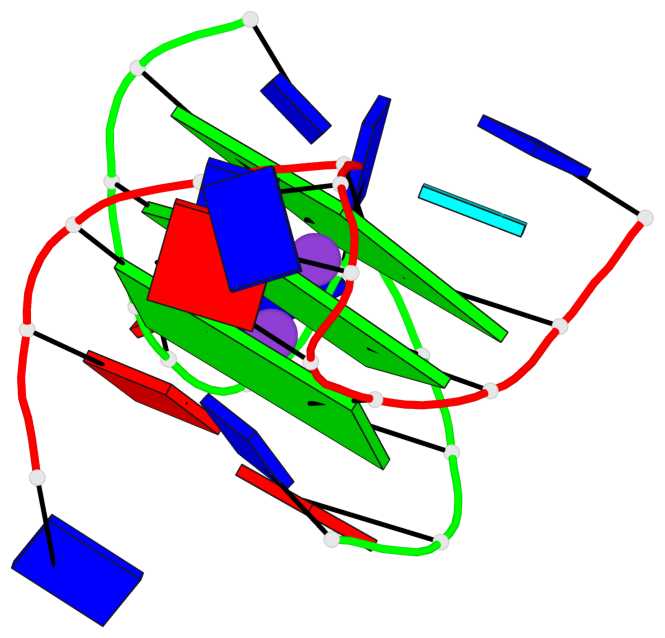
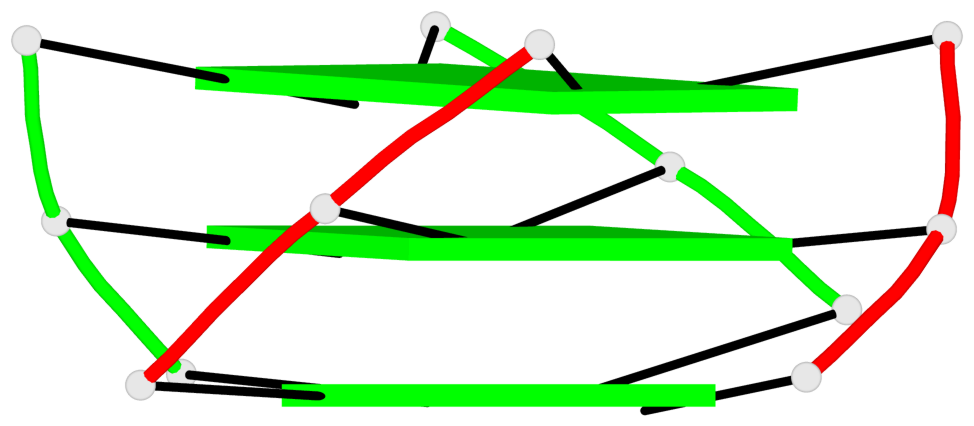
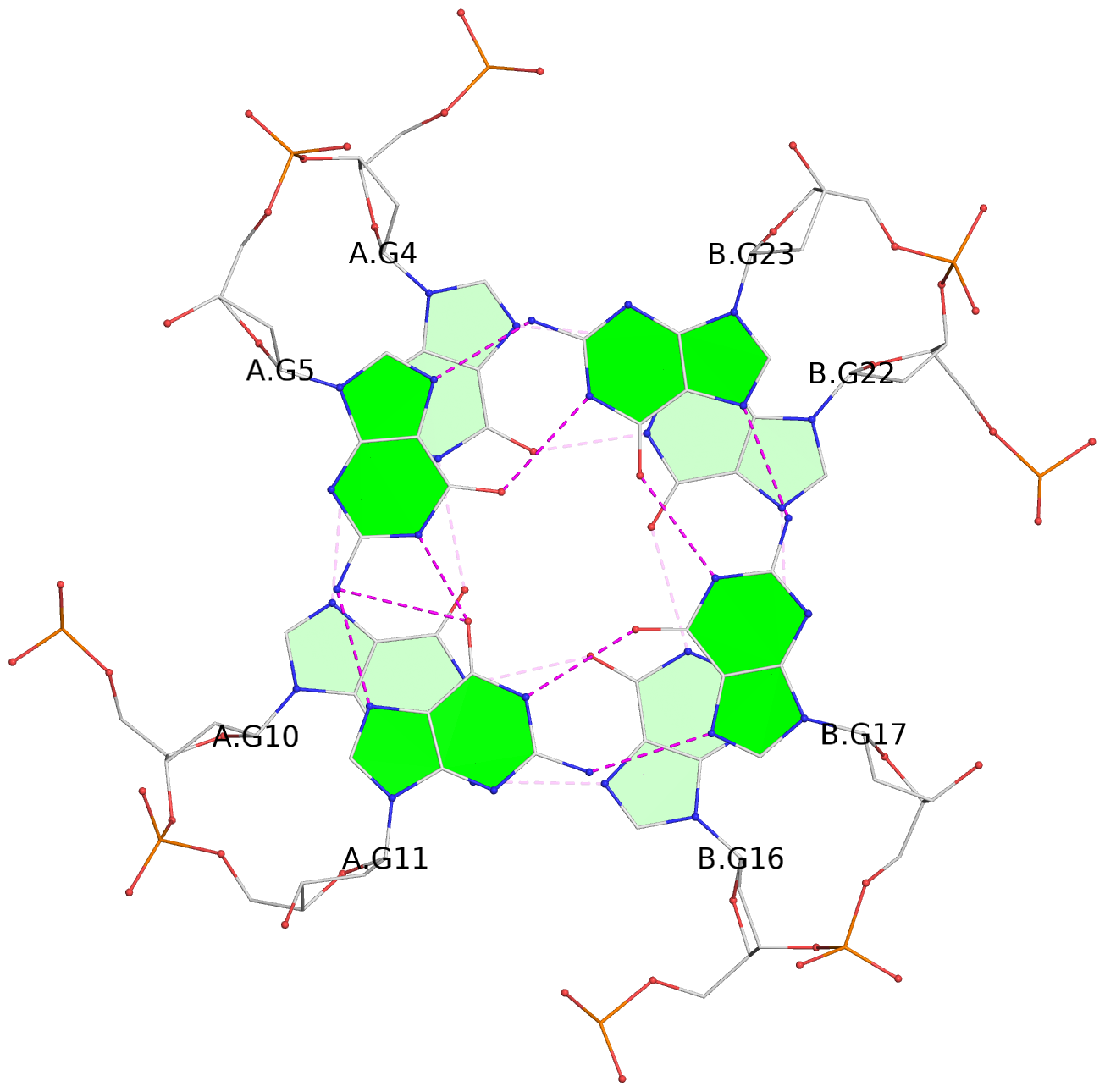
Base-block schematics in six views
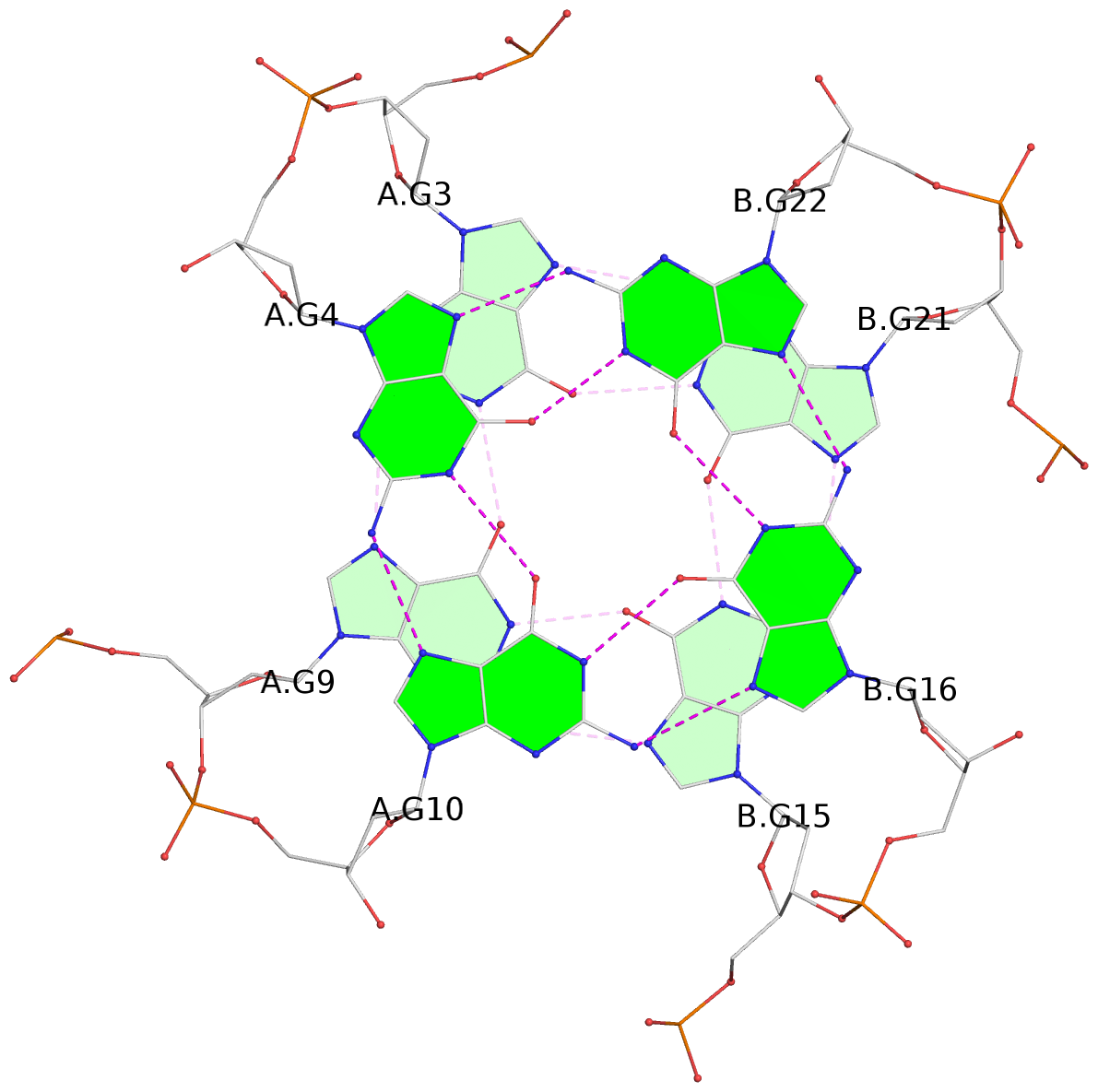
List of 3 G-tetrads
1 glyco-bond=---- sugar=--3- groove=---- planarity=0.154 type=planar nts=4 GGGG A.DG3,A.DG9,B.DG15,B.DG21 2 glyco-bond=---- sugar=---- groove=---- planarity=0.203 type=other nts=4 GGGG A.DG4,A.DG10,B.DG16,B.DG22 3 glyco-bond=---- sugar=-3-- groove=---- planarity=0.253 type=bowl nts=4 GGGG A.DG5,A.DG11,B.DG17,B.DG23
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.