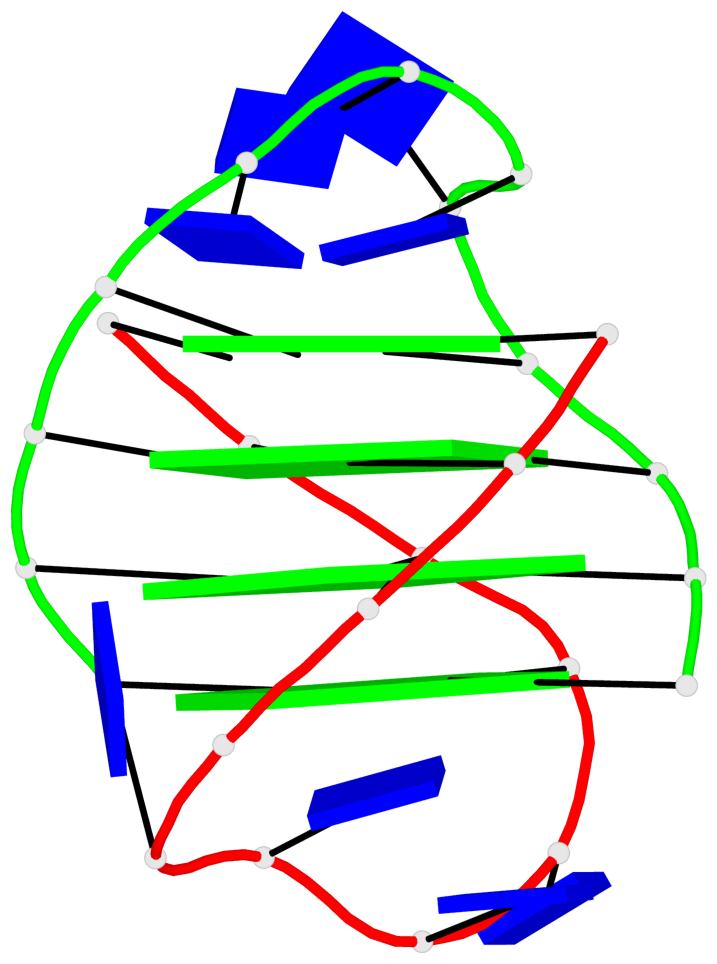
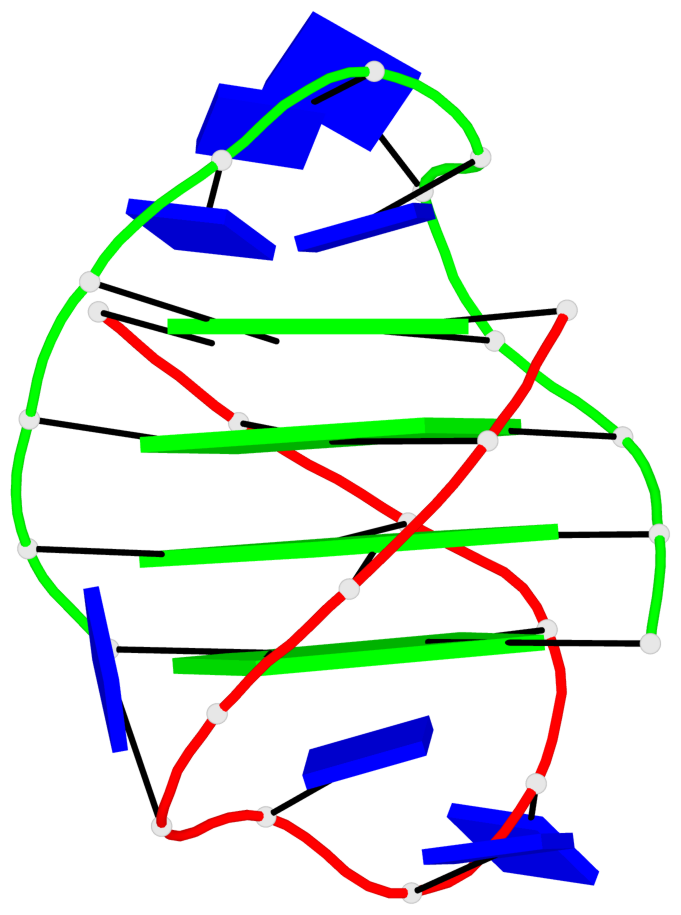
Detailed DSSR results for the G-quadruplex: PDB entry 3eui
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 3eui
- Class
- DNA
- Method
- X-ray (2.2 Å)
- Summary
- A bimolecular anti-parallel-stranded oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine bsu-6042 in a large unit cell
- Reference
- Campbell NH, Patel M, Tofa AB, Ghosh R, Parkinson GN, Neidle S (2009): "Selectivity in Ligand Recognition of G-Quadruplex Loops." Biochemistry, 48, 1675-1680. doi: 10.1021/bi802233v.
- Abstract
- A series of disubstituted acridine ligands have been cocrystallized with a bimolecular DNA G-quadruplex. The ligands have a range of cyclic amino end groups of varying size. The crystal structures show that the diagonal loop in this quadruplex results in a large cavity for these groups, in contrast to the steric constraints imposed by propeller loops in human telomeric quadruplexes. We conclude that the nature of the loop has a significant influence on ligand selectivity for particular quadruplex folds.
- G4 notes
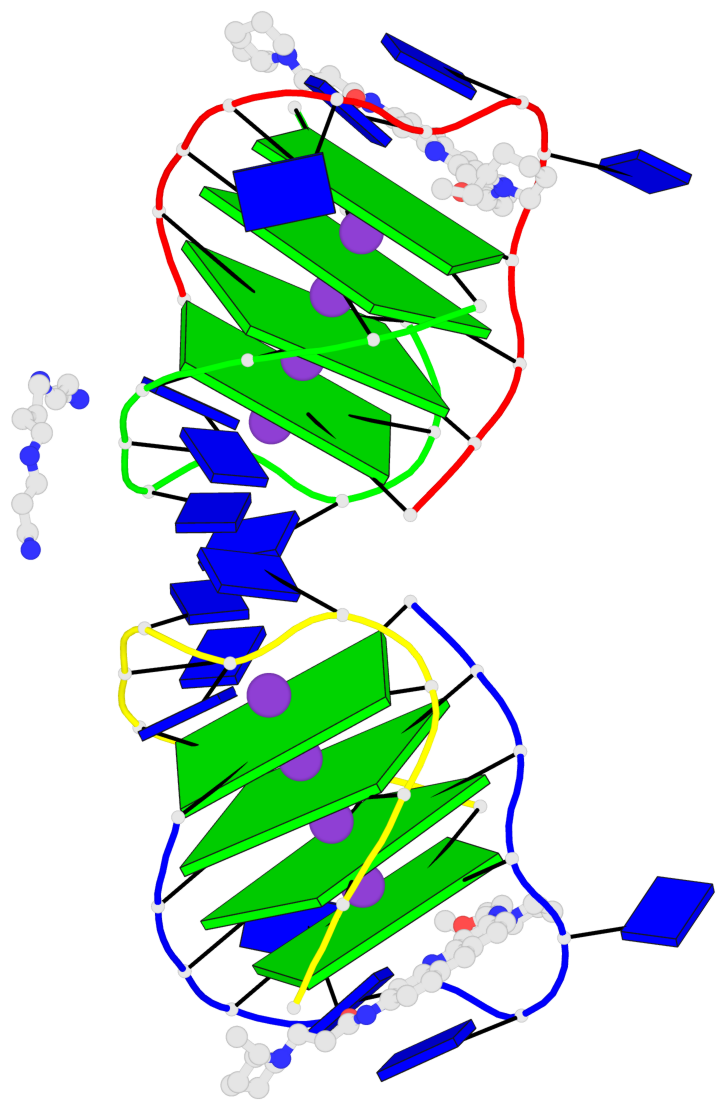
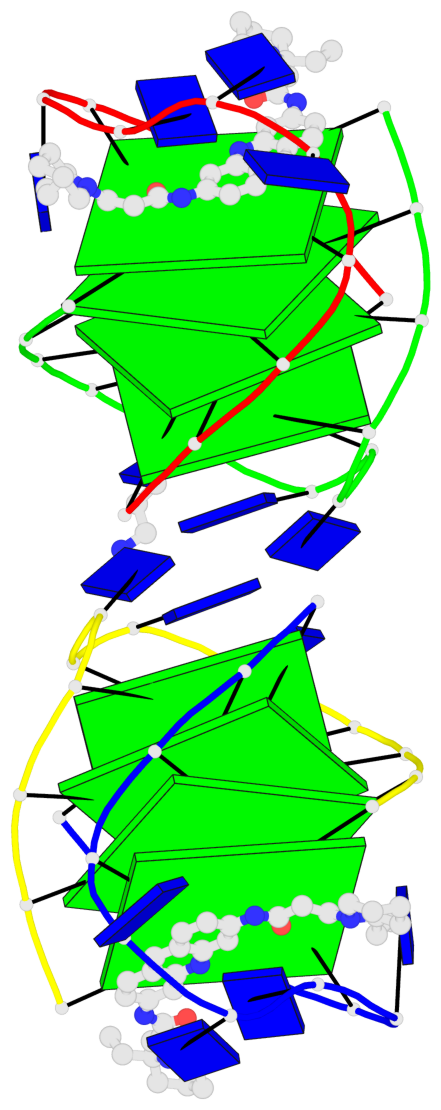
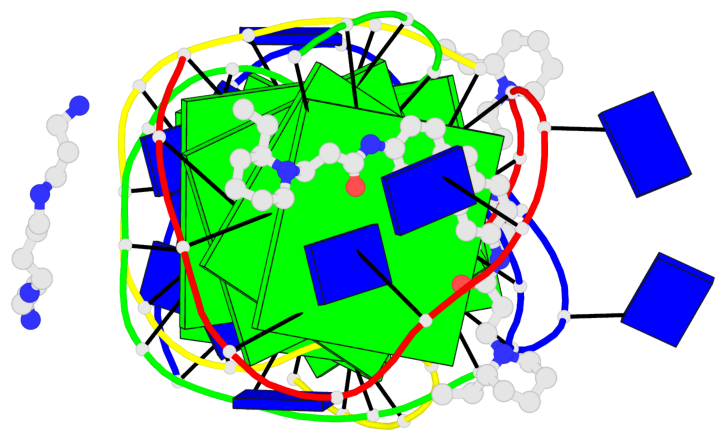
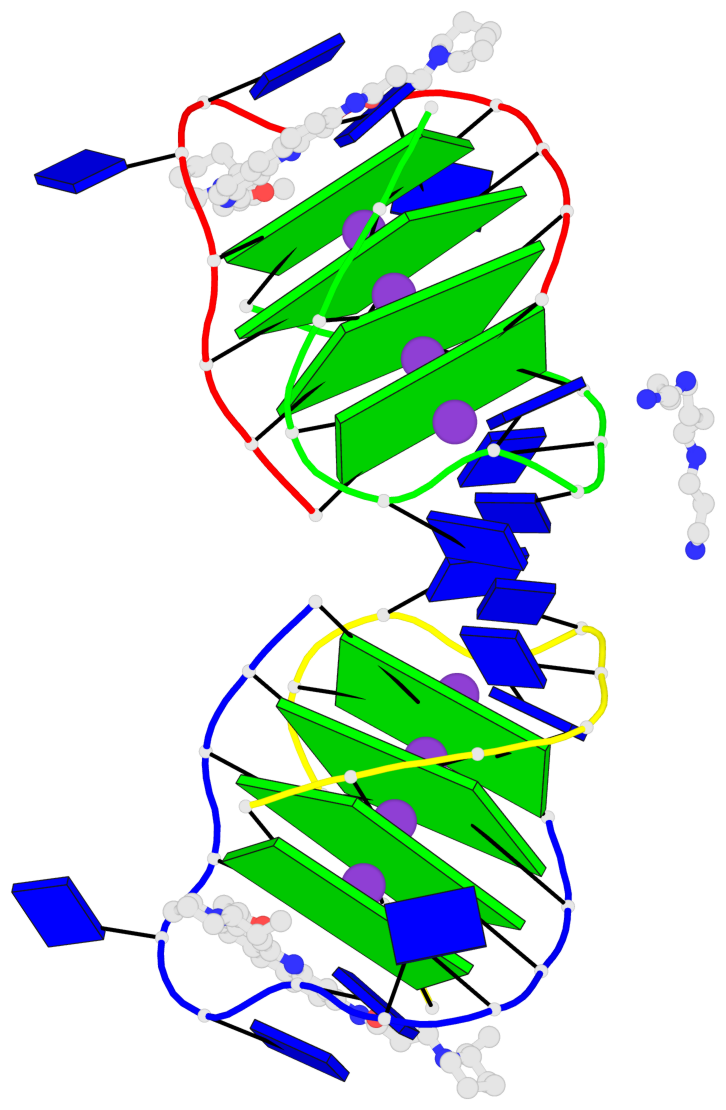
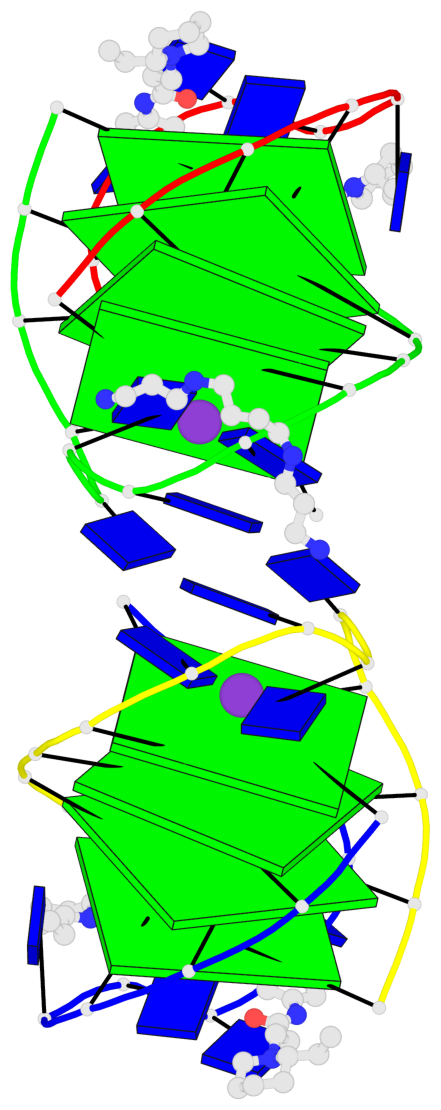
- 8 G-tetrads, 2 G4 helices, 2 G4 stems, (2+2), UDDU
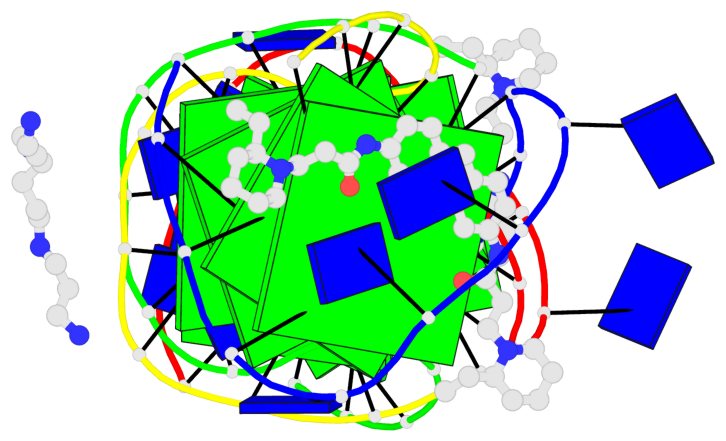
Base-block schematics in six views
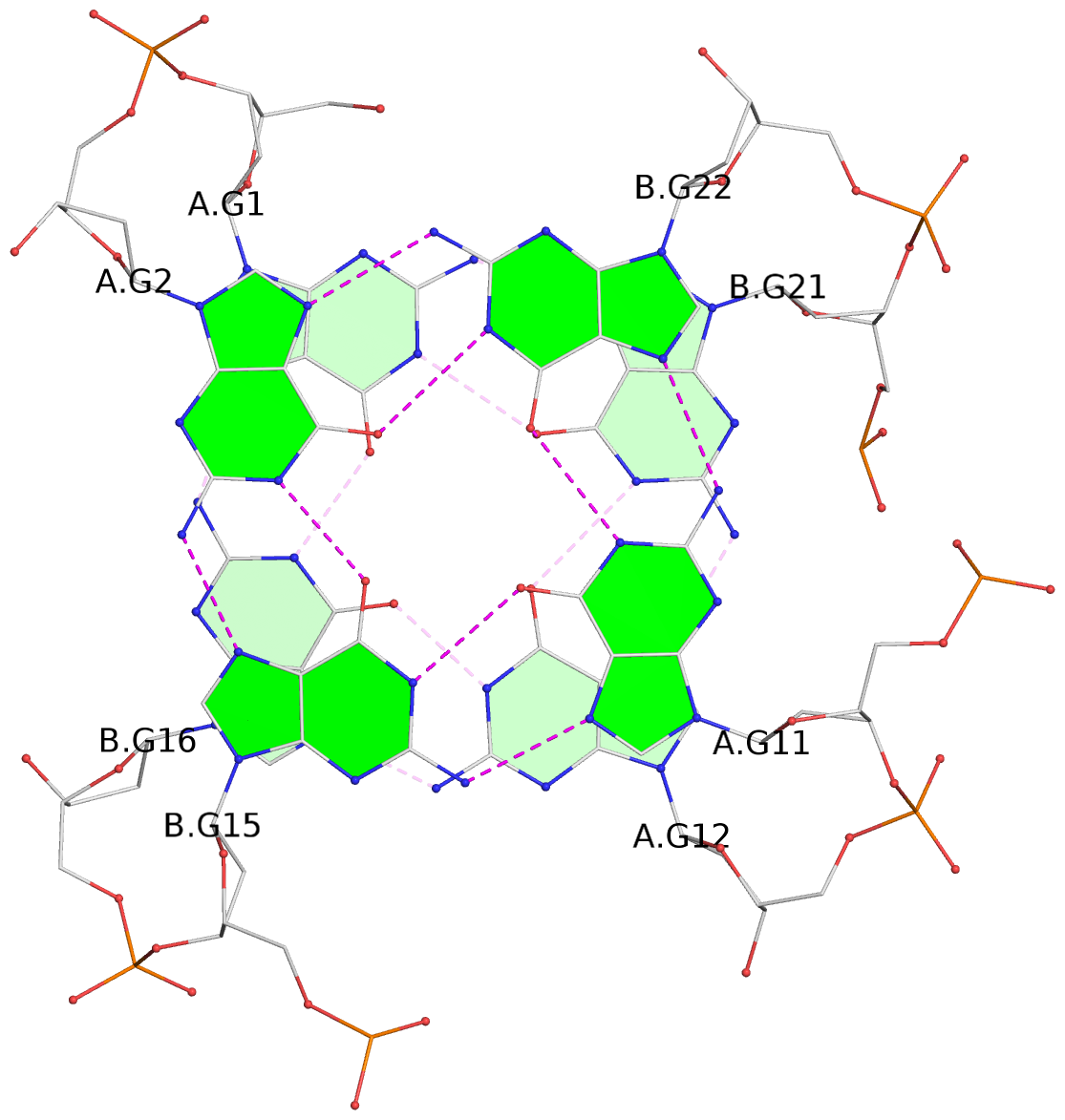
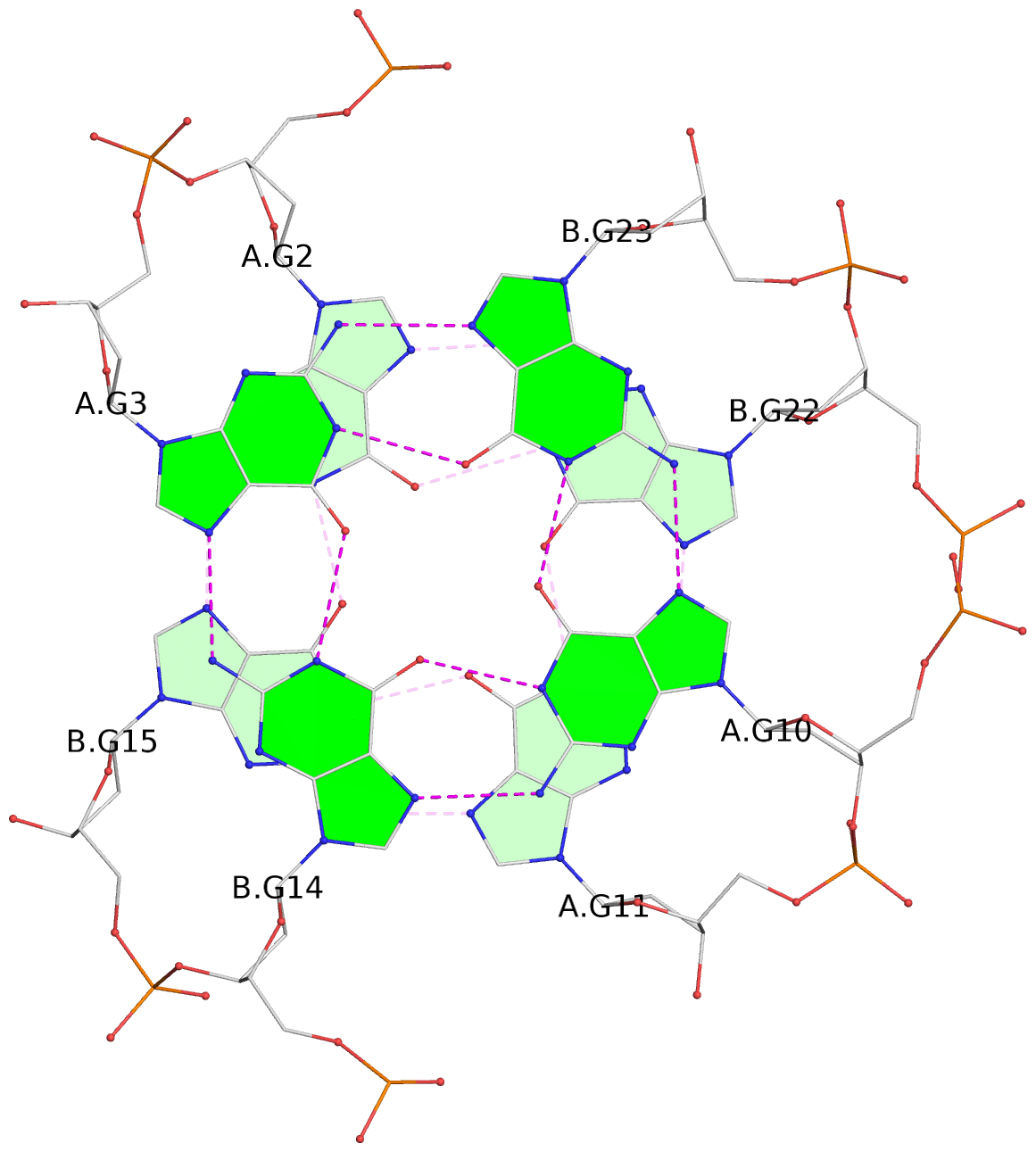
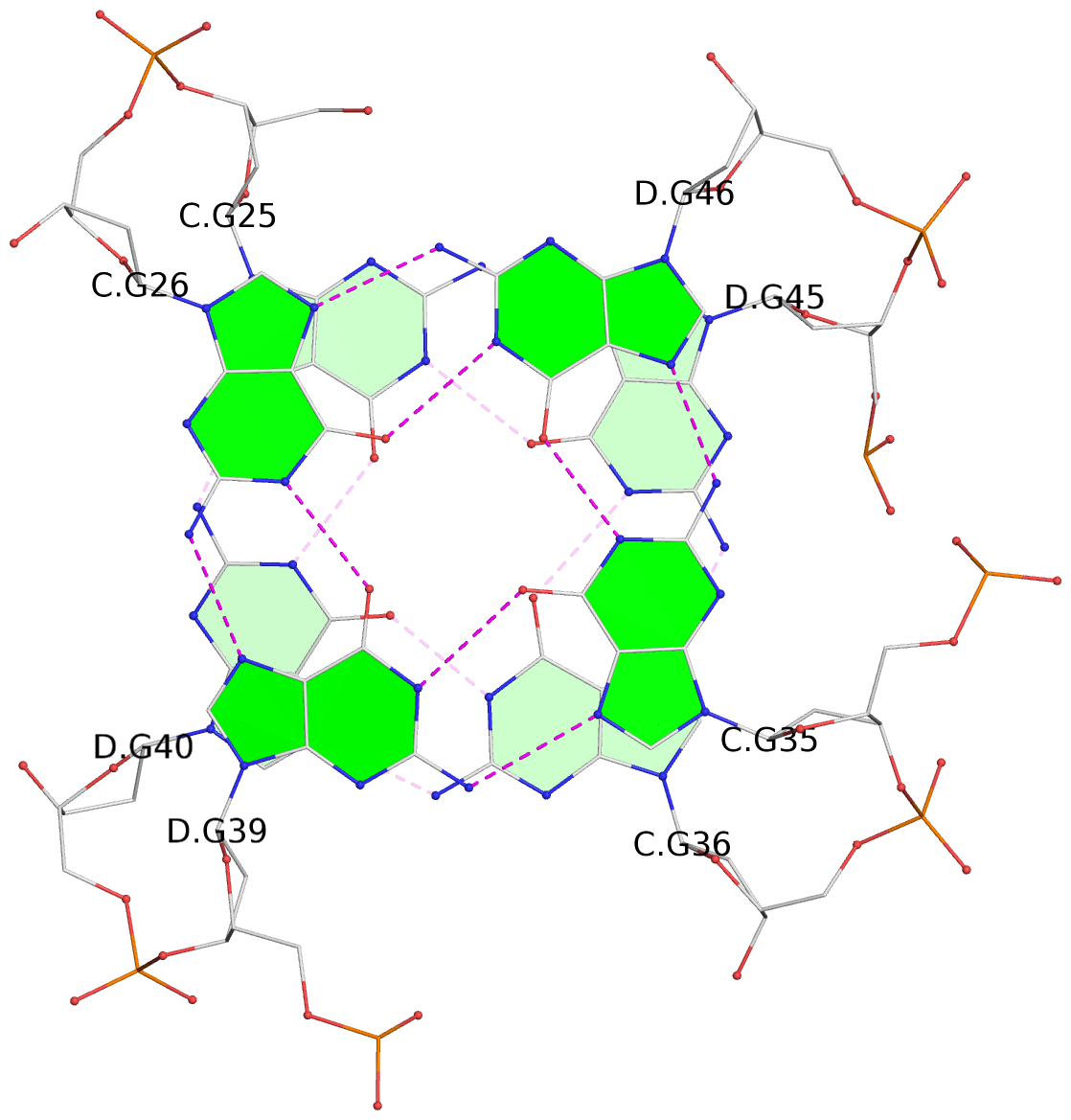
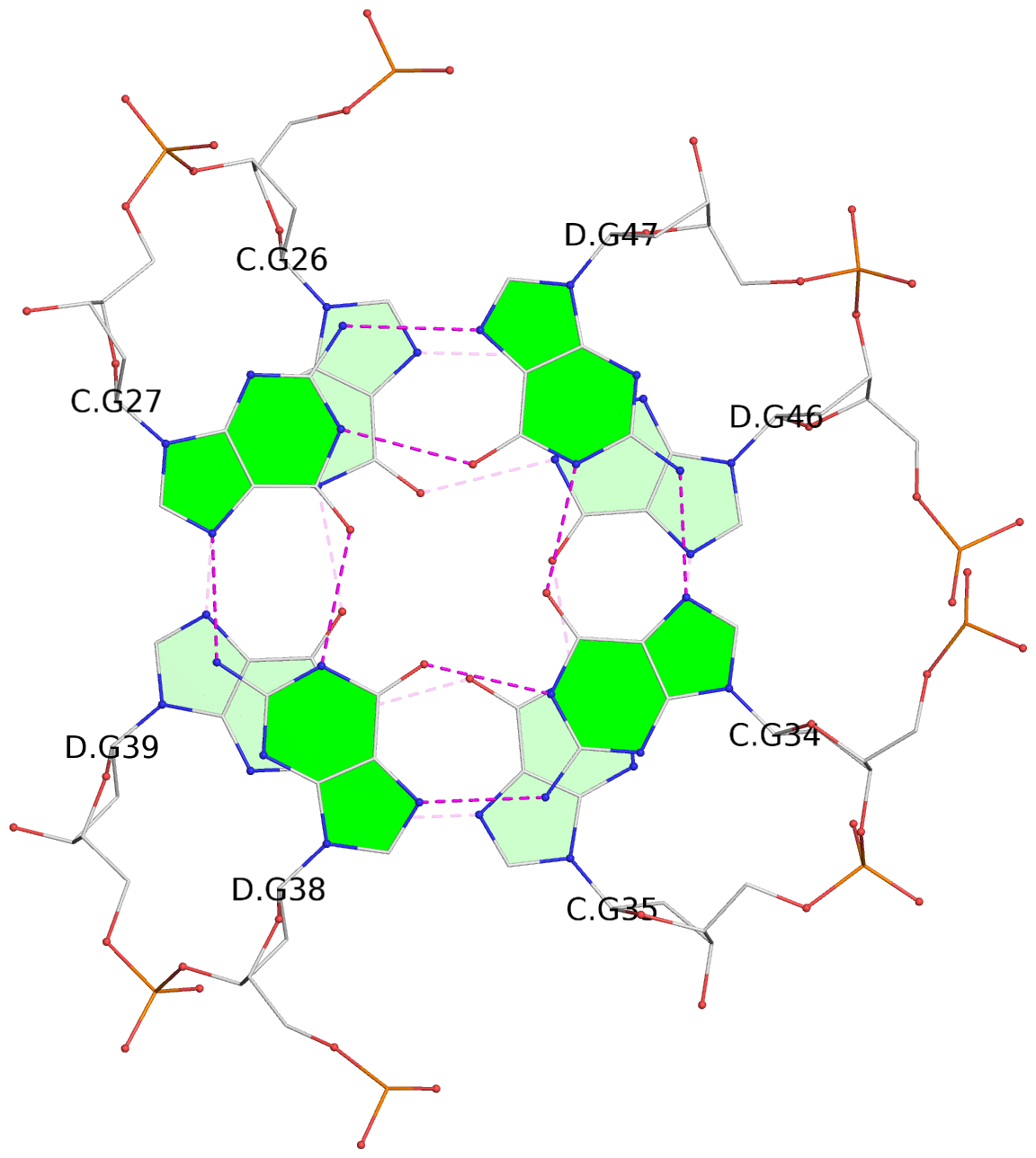
List of 8 G-tetrads
1 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.246 type=bowl nts=4 GGGG A.DG1,B.DG16,A.DG12,B.DG21 2 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.176 type=other nts=4 GGGG A.DG2,B.DG15,A.DG11,B.DG22 3 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.174 type=other nts=4 GGGG A.DG3,B.DG14,A.DG10,B.DG23 4 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.190 type=other nts=4 GGGG A.DG4,B.DG13,A.DG9,B.DG24 5 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.277 type=bowl nts=4 GGGG C.DG25,D.DG40,C.DG36,D.DG45 6 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.209 type=other nts=4 GGGG C.DG26,D.DG39,C.DG35,D.DG46 7 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.179 type=other nts=4 GGGG C.DG27,D.DG38,C.DG34,D.DG47 8 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.211 type=other nts=4 GGGG C.DG28,D.DG37,C.DG33,D.DG48
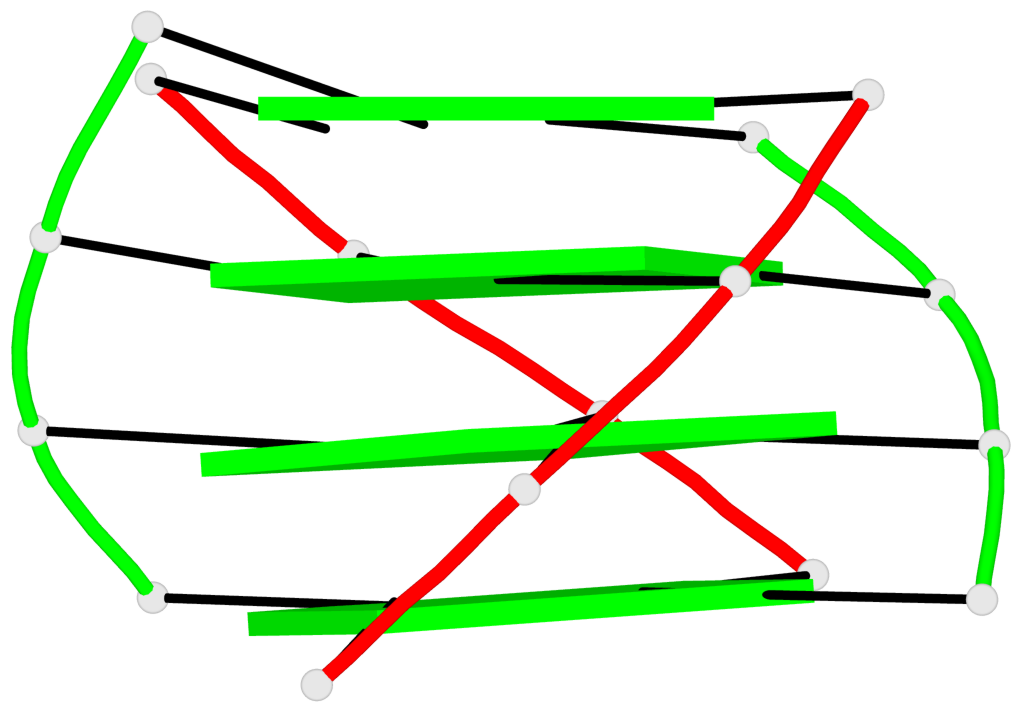
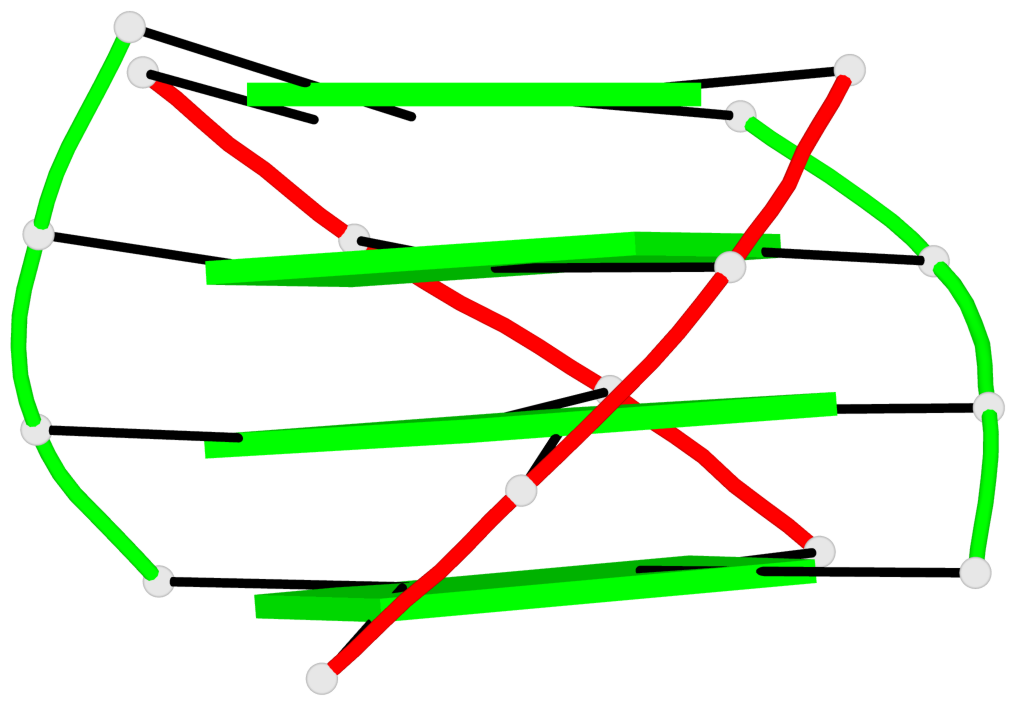
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 1 stem
Helix#2, 4 G-tetrad layers, inter-molecular, with 1 stem
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.