Detailed DSSR results for the G-quadruplex: PDB entry 3nz7
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 3nz7
- Class
- DNA
- Method
- X-ray (1.1 Å)
- Summary
- A bimolecular anti-parallel-stranded oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine ligand containing bis-3-fluoropyrrolidine end side chains
- Reference
- Campbell NH, Smith DL, Reszka AP, Neidle S, O'Hagan D (2011): "Fluorine in medicinal chemistry: beta-fluorination of peripheral pyrrolidines attached to acridine ligands affects their interactions with G-quadruplex DNA." Org.Biomol.Chem., 9, 1328-1331. doi: 10.1039/c0ob00886a.
- Abstract
- Comparative X-ray structure studies reveal that C-F bond incorporation into the peripheral pyrrolidine moieties of the G-quadruplex DNA binding ligand BSU6039 leads to a distinct pyrrolidine ring conformation, relative to the non-fluorinated analogue, and with a different binding mode involving reversal of the pyrrolidinium N(+)-H orientation.
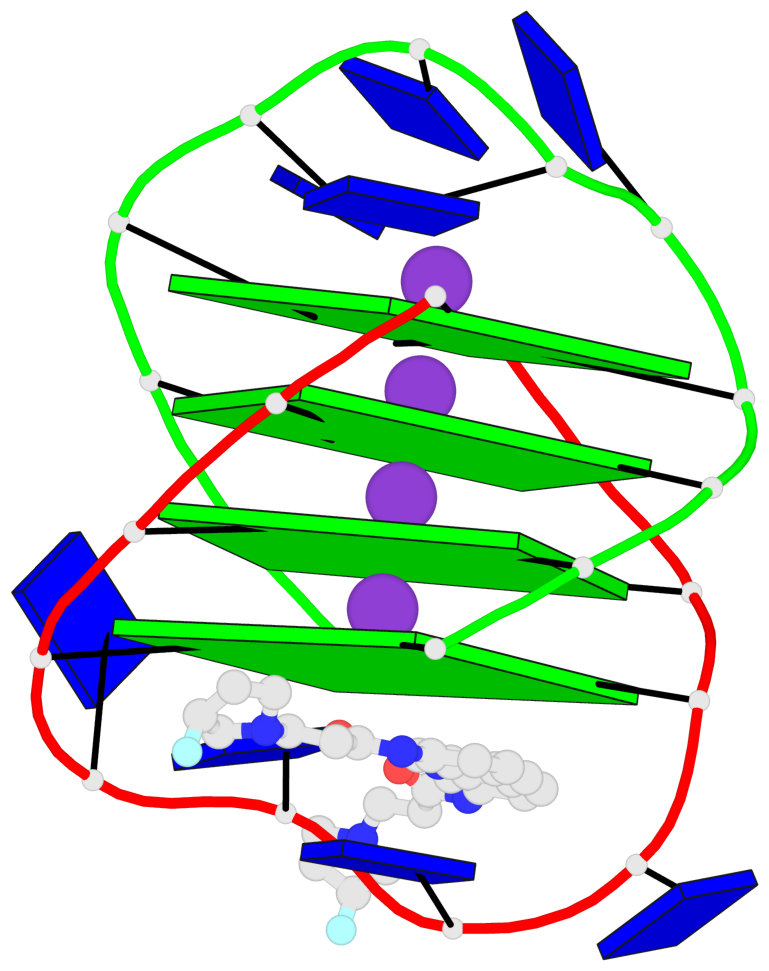
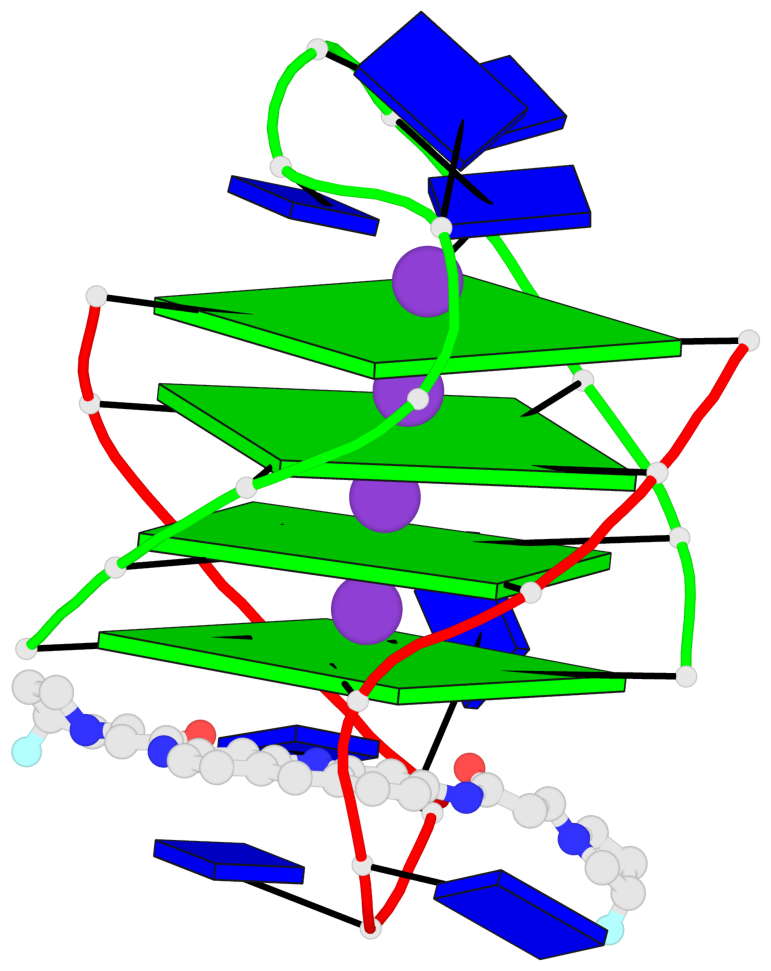
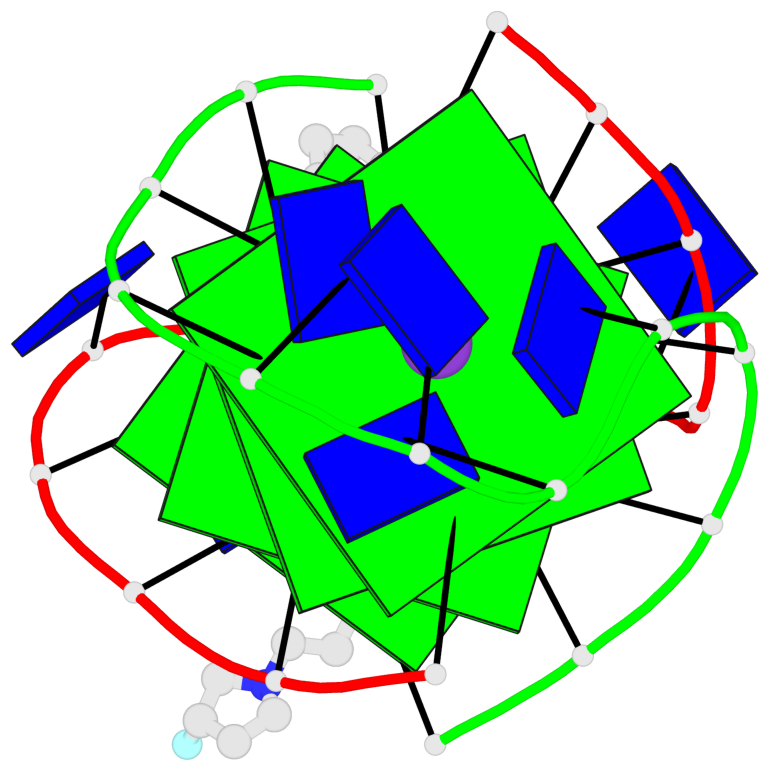
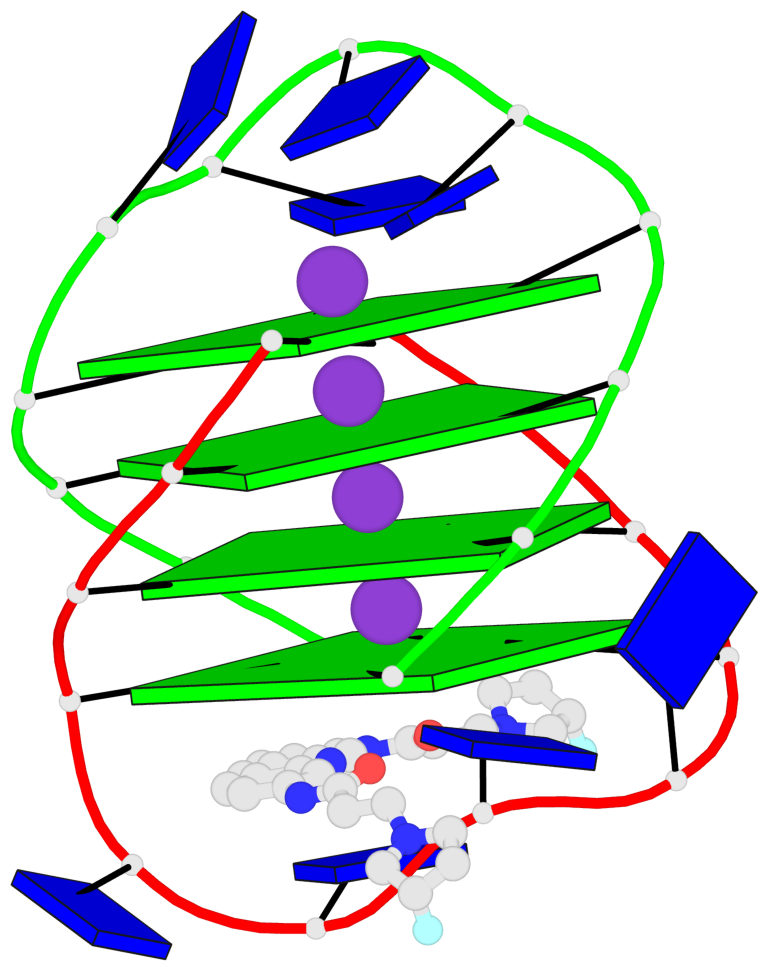
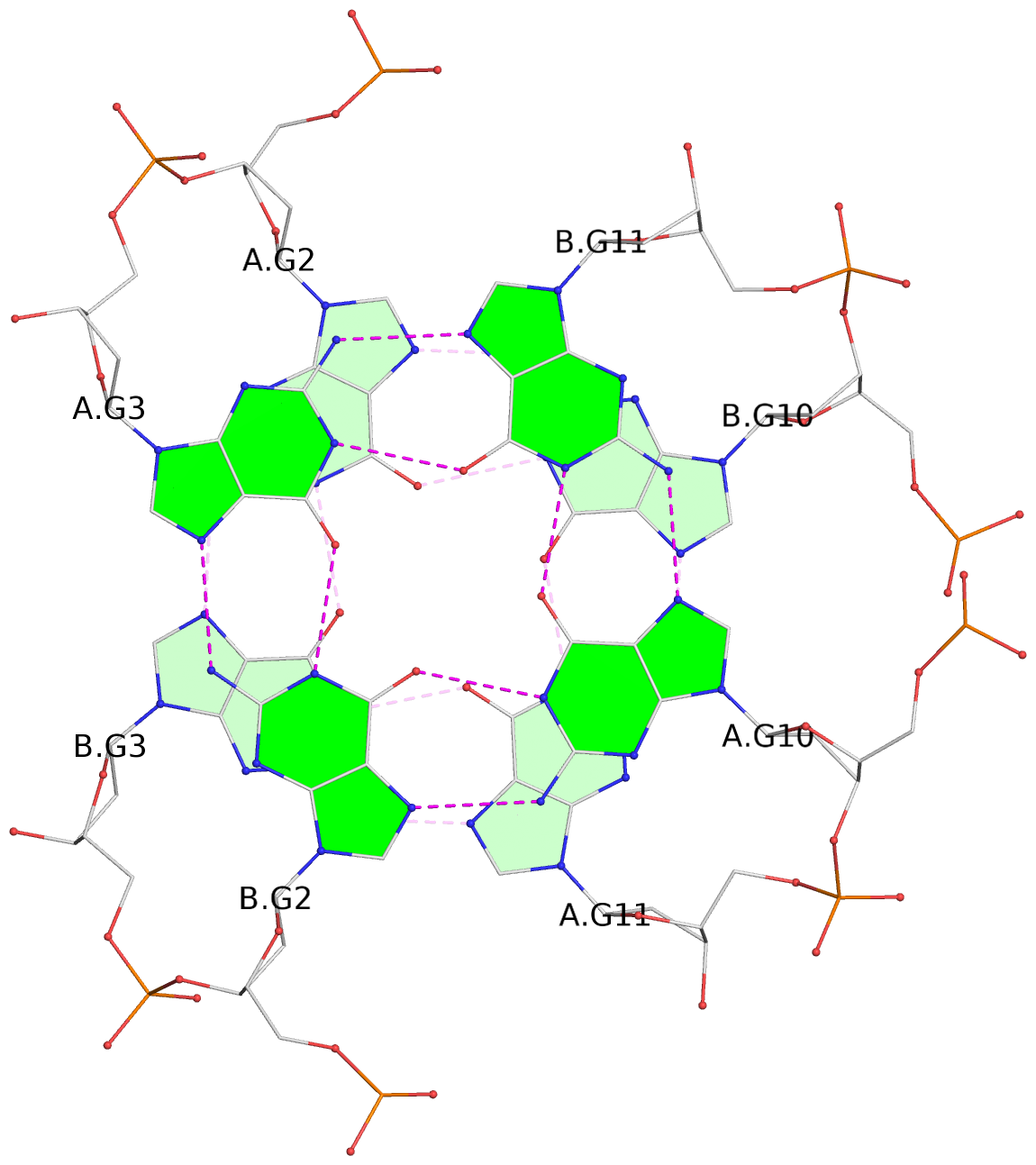
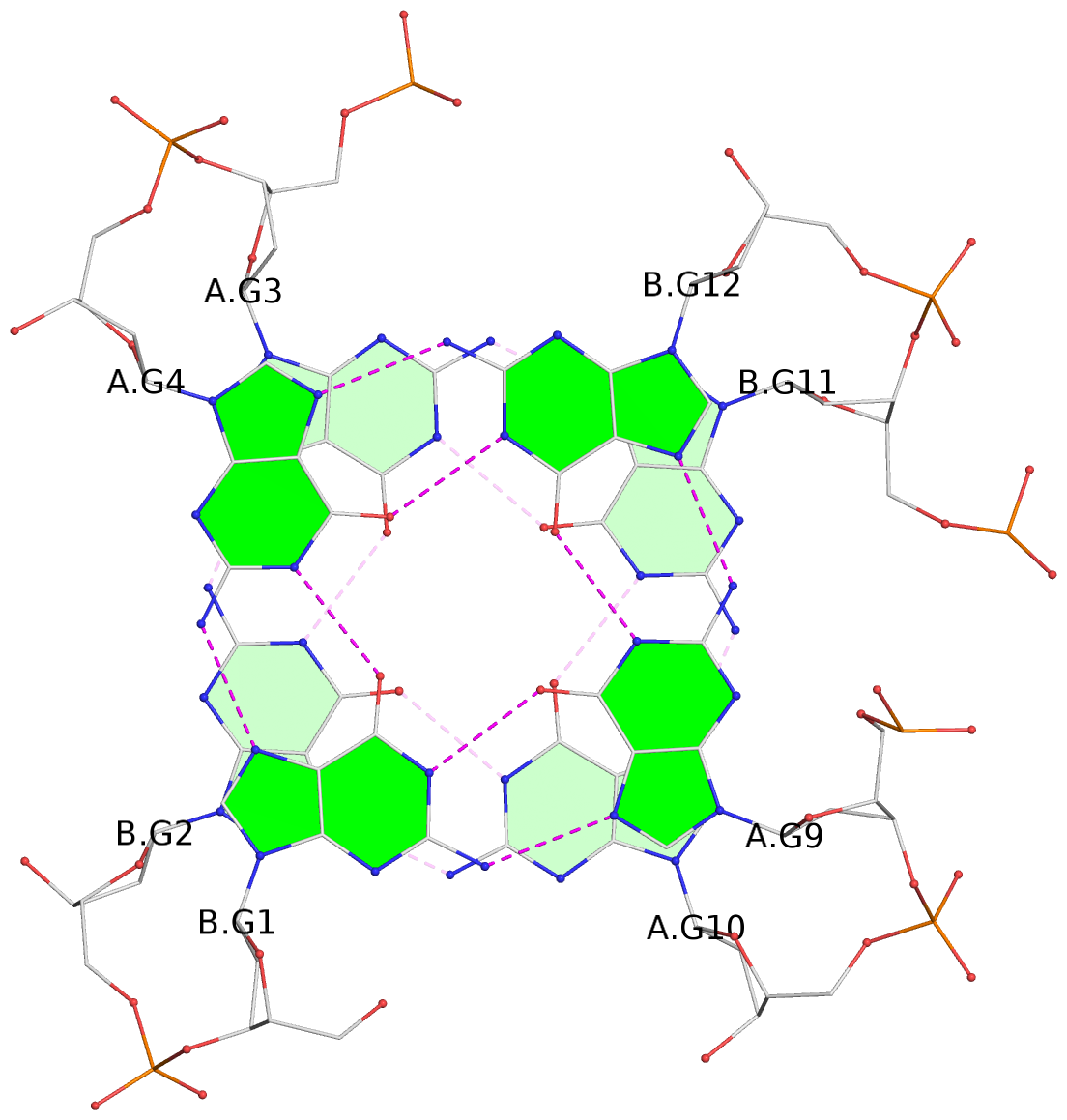
- G4 notes
- 4 G-tetrads, 1 G4 helix, 1 G4 stem, (2+2), UDDU
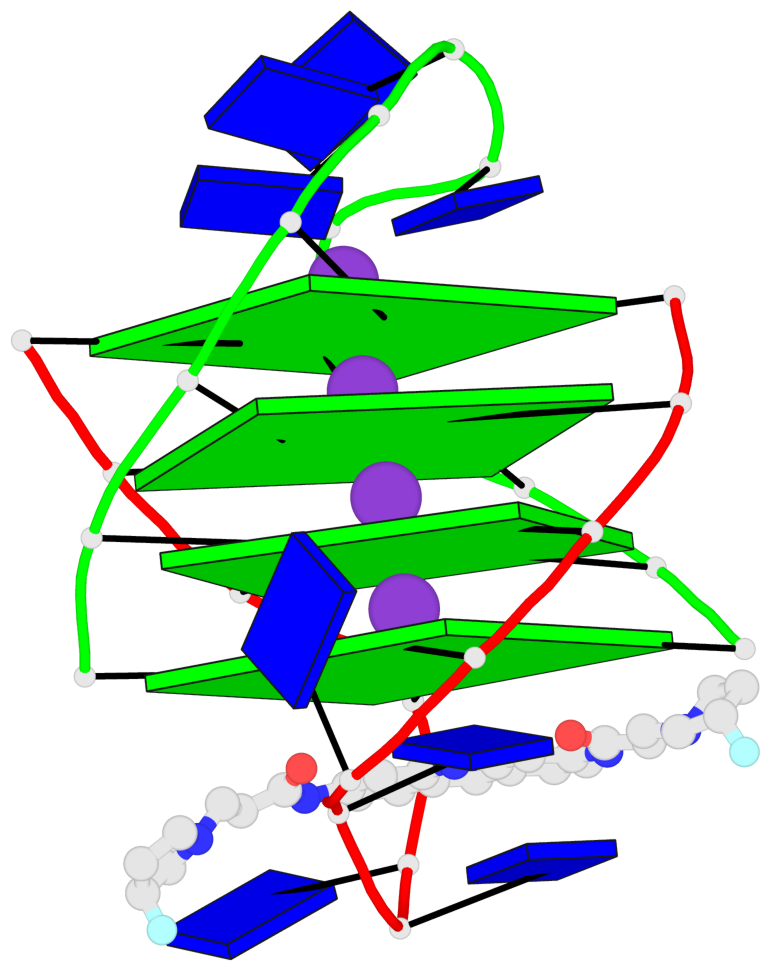
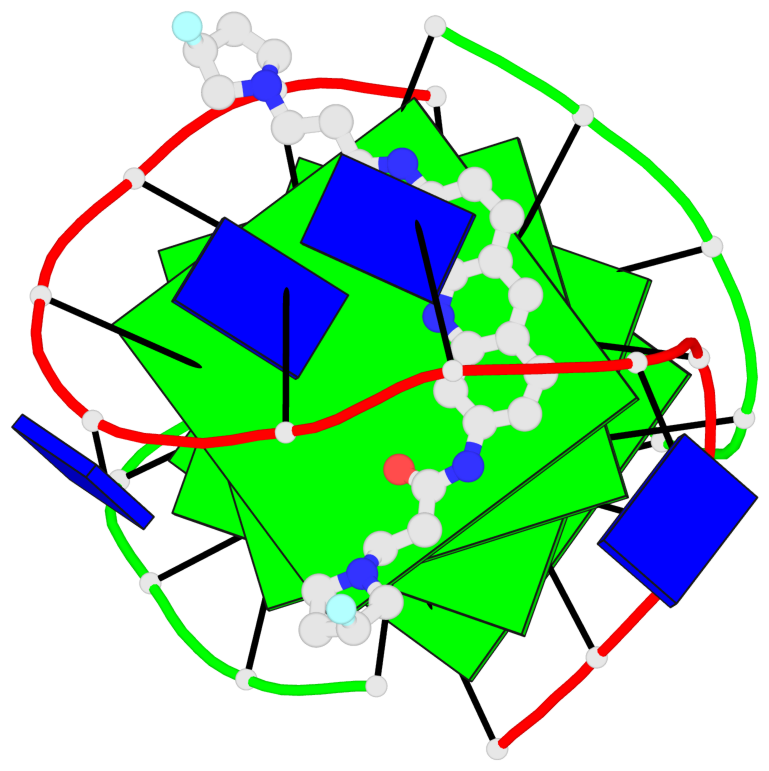
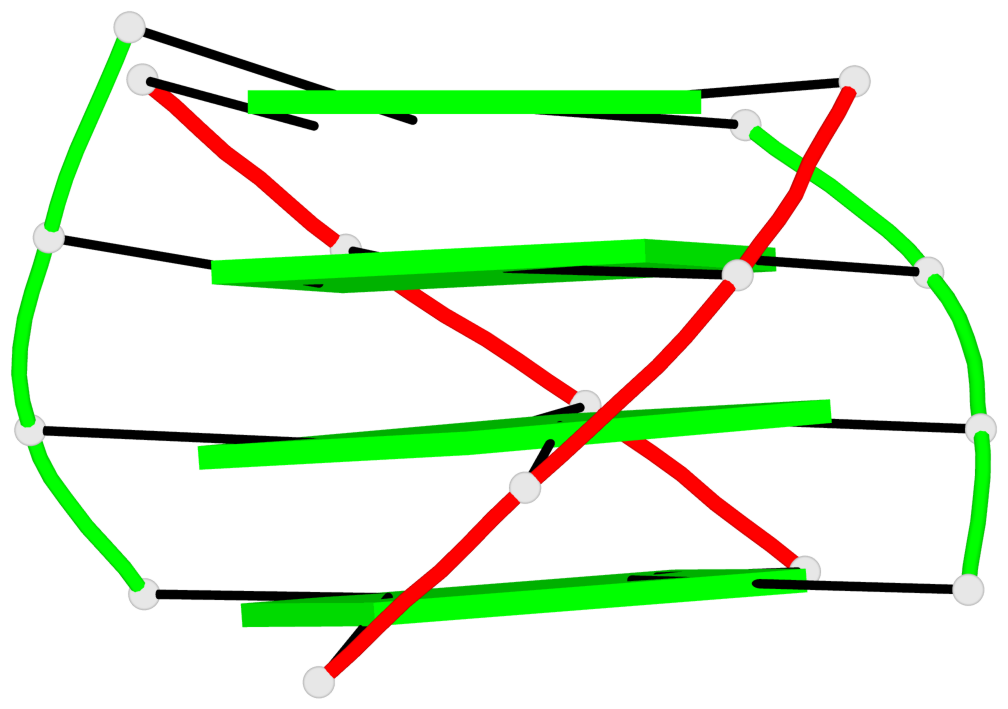
Base-block schematics in six views
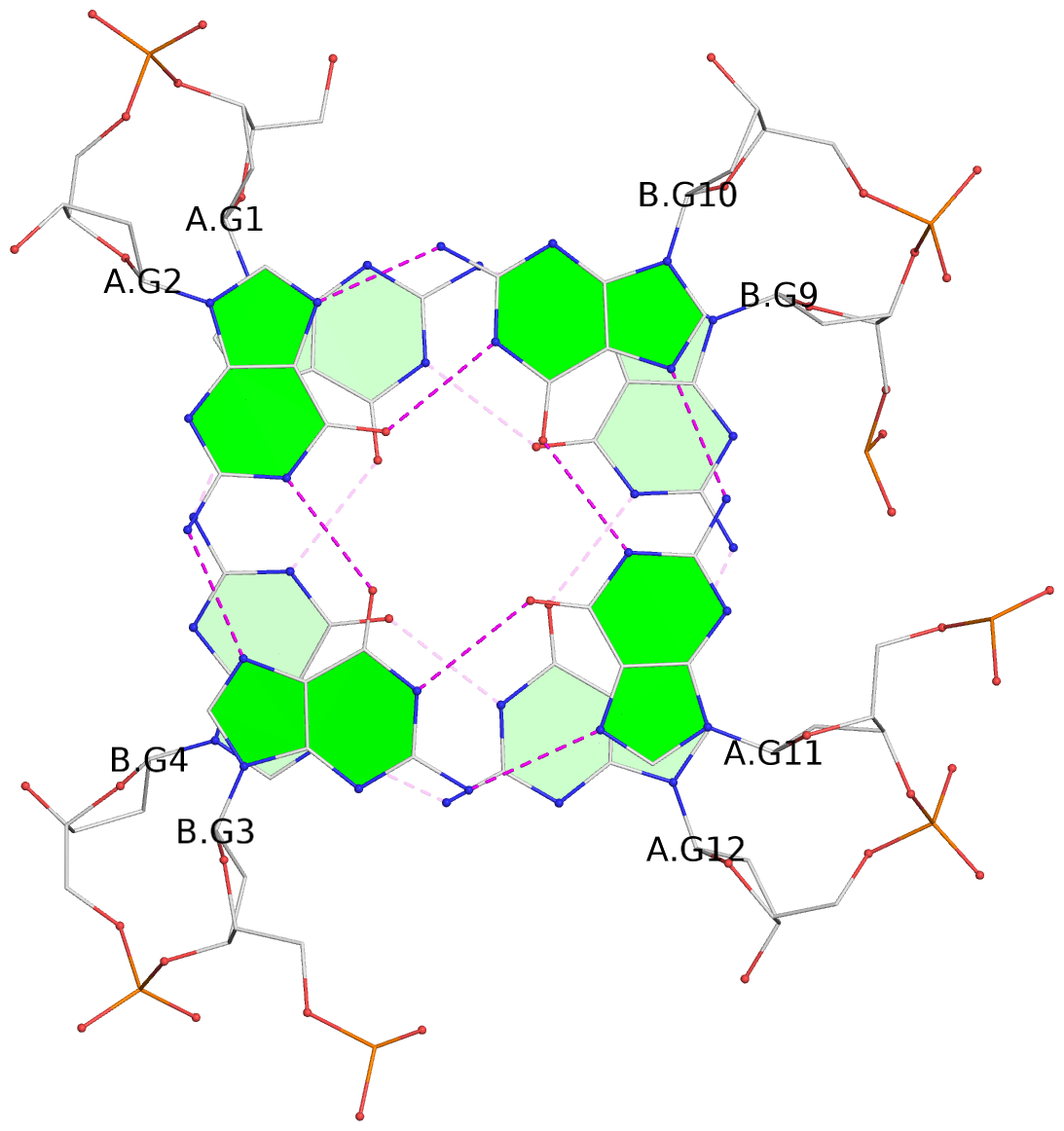
List of 4 G-tetrads
1 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.263 type=bowl nts=4 GGGG A.DG1,B.DG4,A.DG12,B.DG9 2 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.185 type=other nts=4 GGGG A.DG2,B.DG3,A.DG11,B.DG10 3 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.173 type=other nts=4 GGGG A.DG3,B.DG2,A.DG10,B.DG11 4 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.204 type=other nts=4 GGGG A.DG4,B.DG1,A.DG9,B.DG12
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.