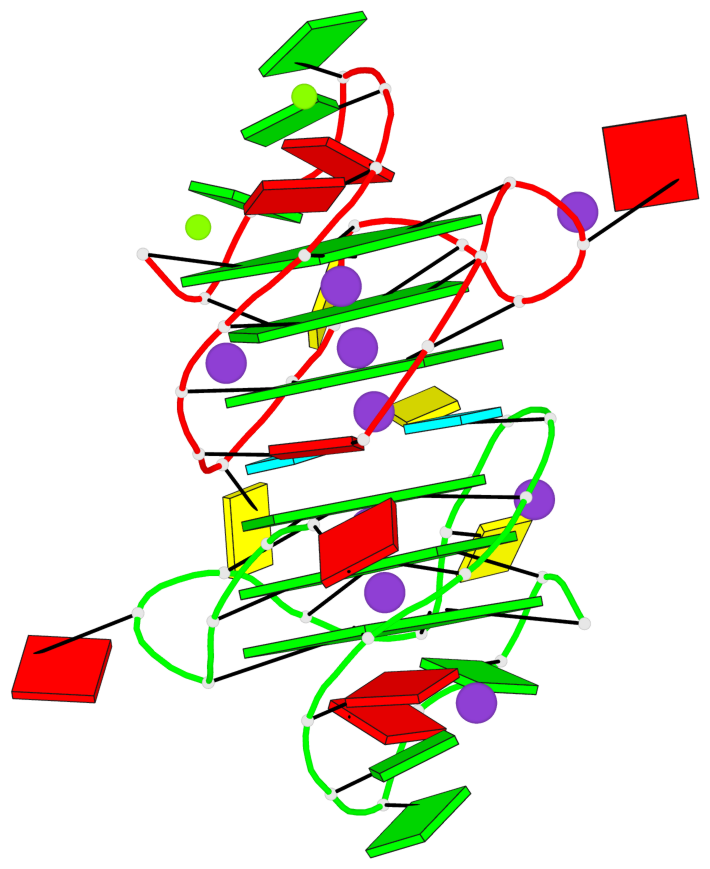
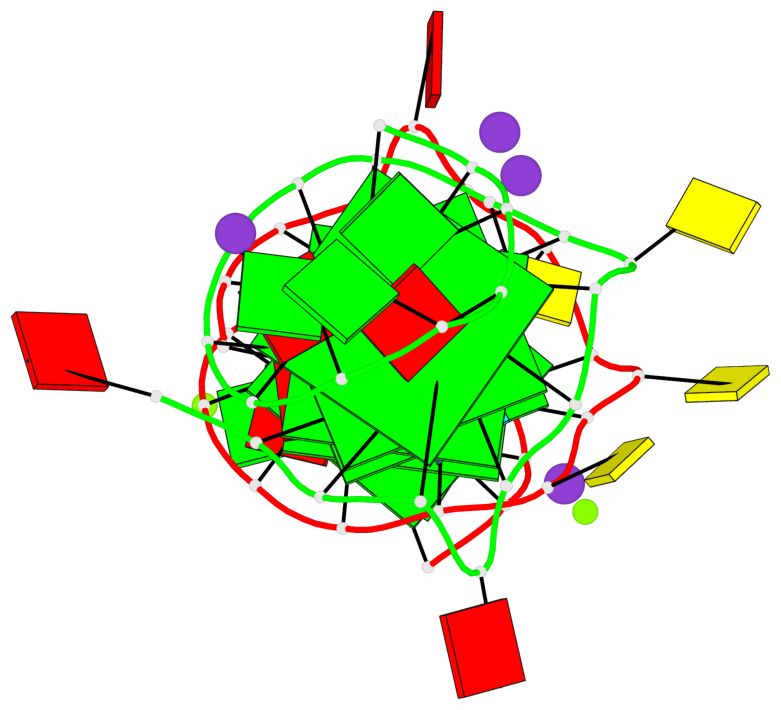
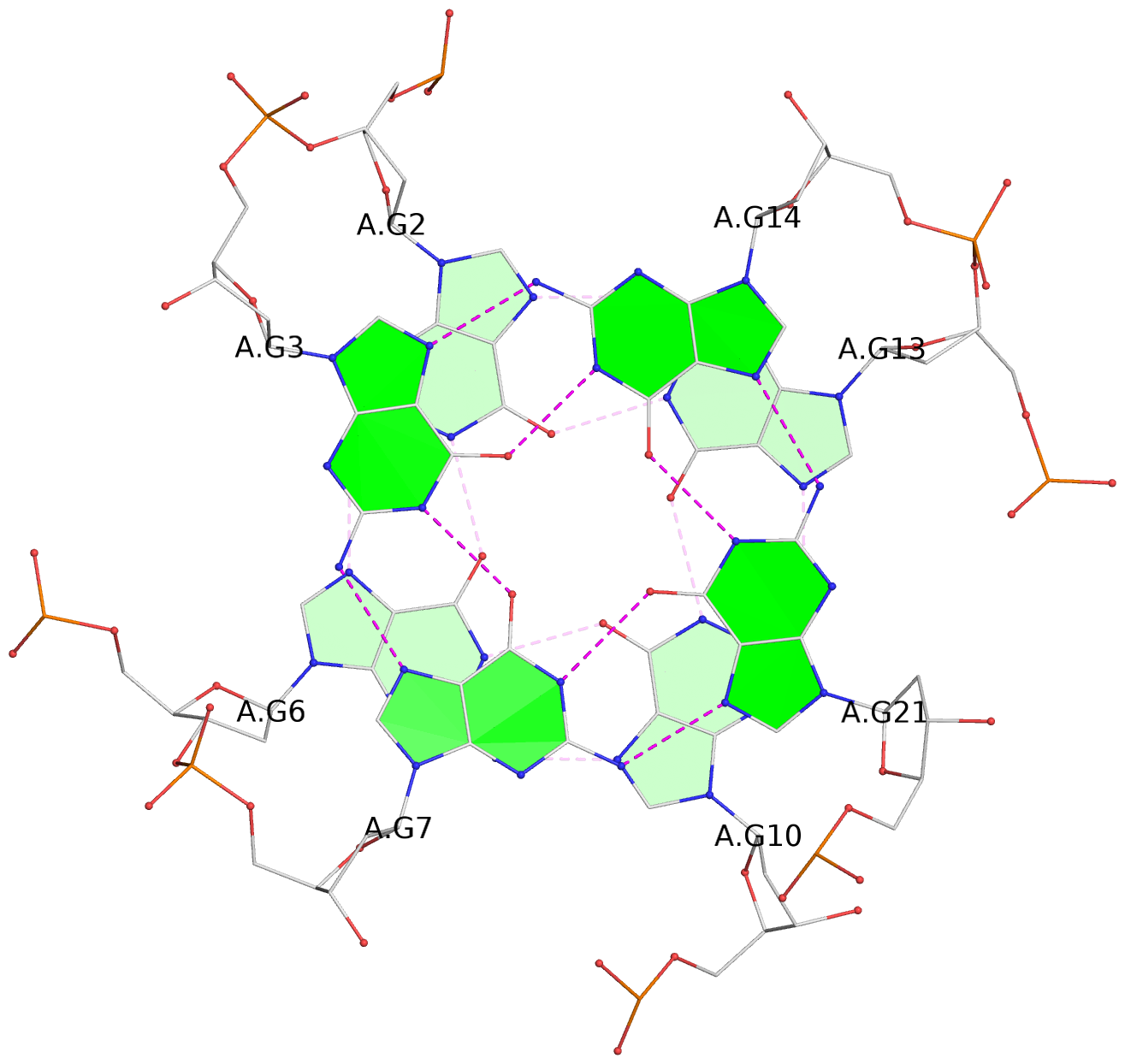
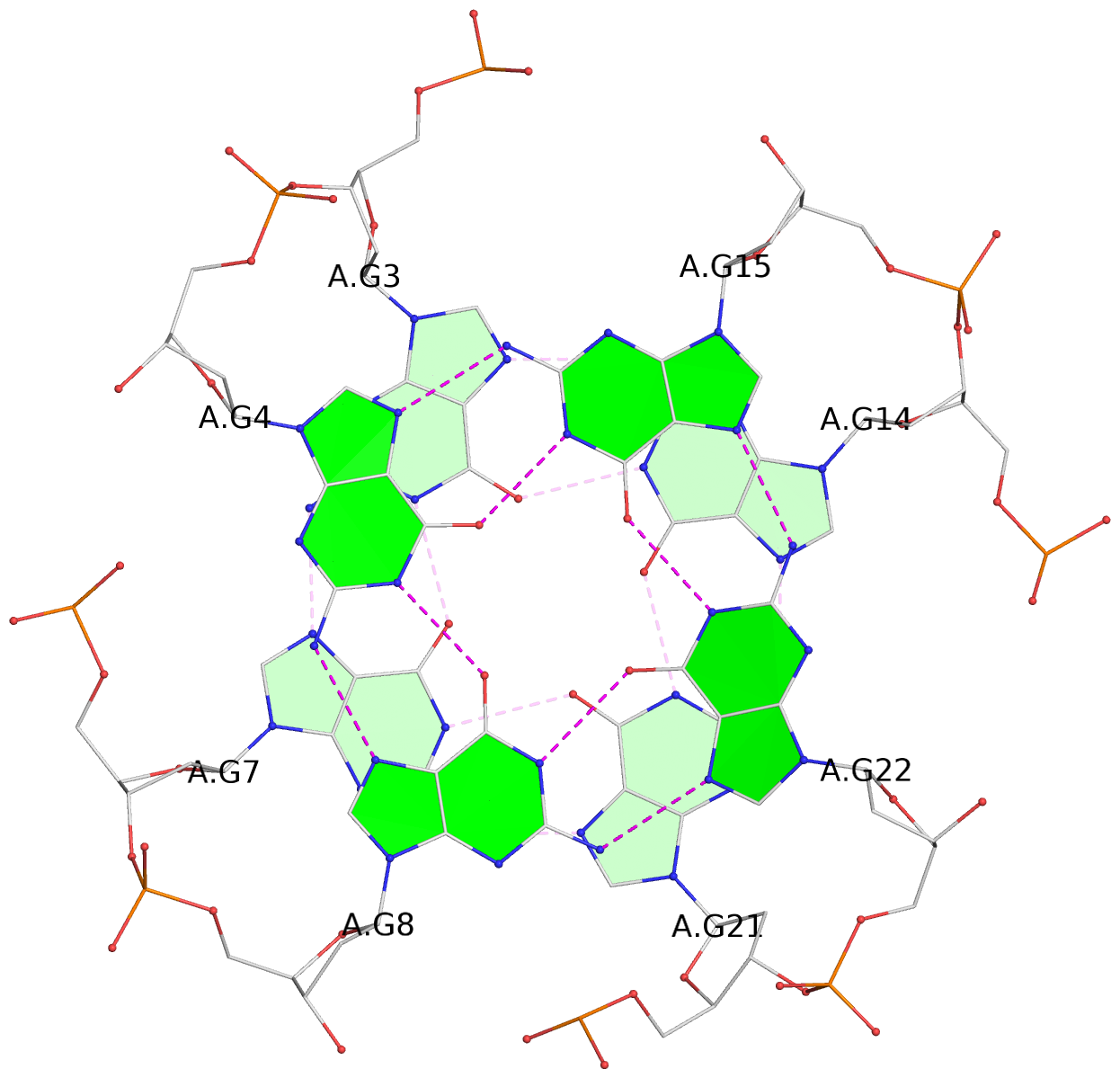
Detailed DSSR results for the G-quadruplex: PDB entry 3qxr
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 3qxr
- Class
- DNA
- Method
- X-ray (1.62 Å)
- Summary
- Crystal structure of the brominated ckit-1 proto-oncogene promoter quadruplex DNA
- Reference
- Wei D, Parkinson GN, Reszka AP, Neidle S (2012): "Crystal structure of a c-kit promoter quadruplex reveals the structural role of metal ions and water molecules in maintaining loop conformation." Nucleic Acids Res., 40, 4691-4700. doi: 10.1093/nar/gks023.
- Abstract
- We report here the 1.62 Å crystal structure of an intramolecular quadruplex DNA formed from a sequence in the promoter region of the c-kit gene. This is the first reported crystal structure of a promoter quadruplex and the first observation of localized magnesium ions in a quadruplex structure. The structure reveals that potassium and magnesium ions have an unexpected yet significant structural role in stabilizing particular quadruplex loops and grooves that is distinct from but in addition to the role of potassium ions in the ion channel at the centre of all quadruplex structures. The analysis also shows how ions cluster together with structured water molecules to stabilize the quadruplex arrangement. This particular quadruplex has been previously studied by NMR methods, and the present X-ray structure is in accord with the earlier topology assignment. However, as well as the observations of potassium and magnesium ions, the crystal structure has revealed a highly significant difference in the dimensions of the large cleft in the structure, which is a plausible target for small molecules. This difference can be understood by the stabilizing role of structured water networks.
- G4 notes
- 6 G-tetrads, 2 G4 helices, 2 G4 stems, 2(-PX+P), parallel(4+0), UUUU
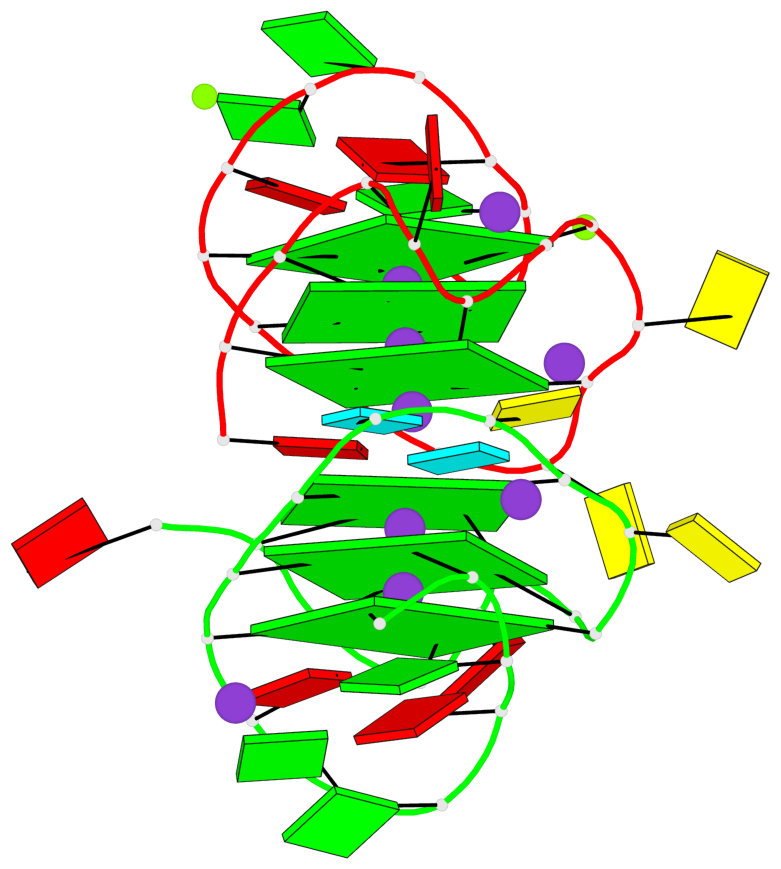
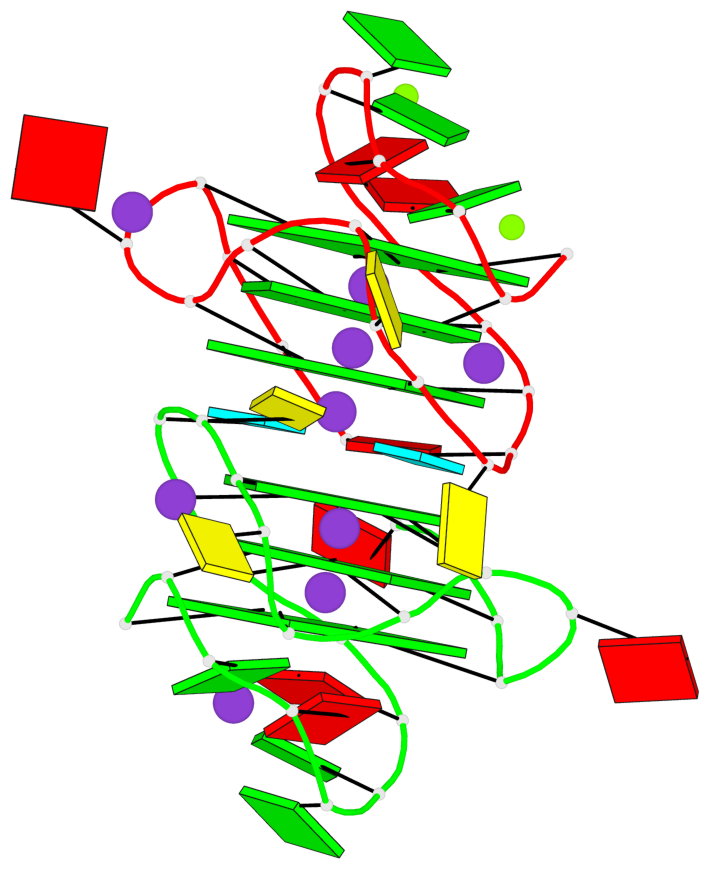
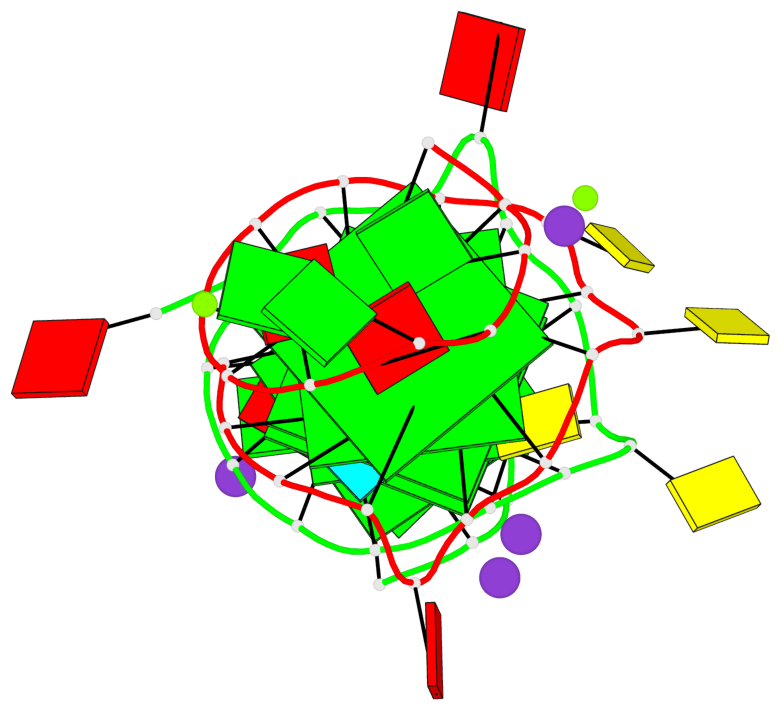
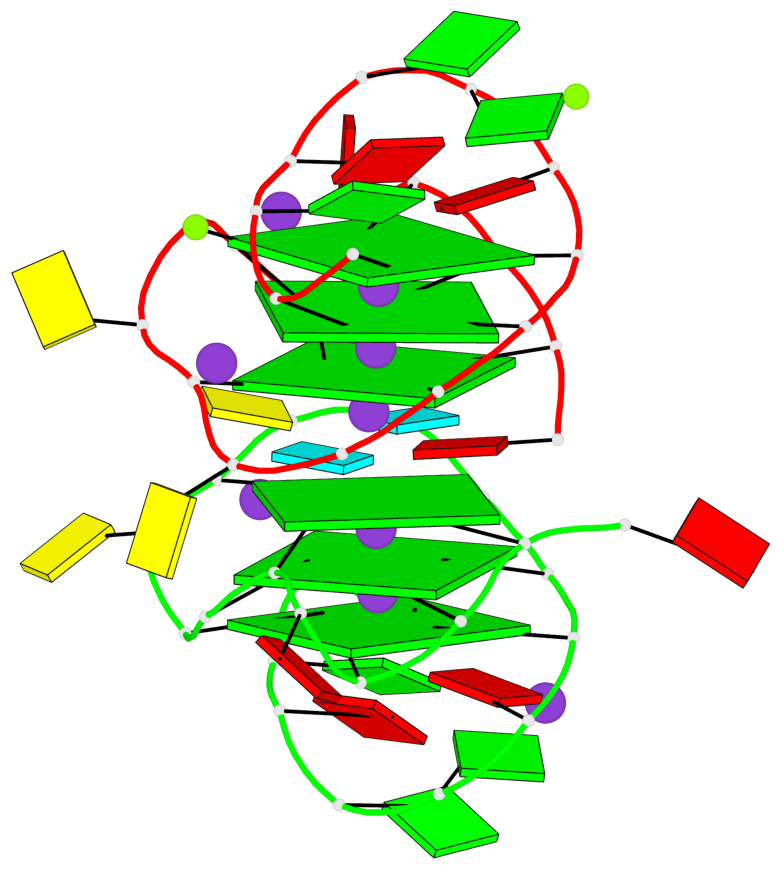
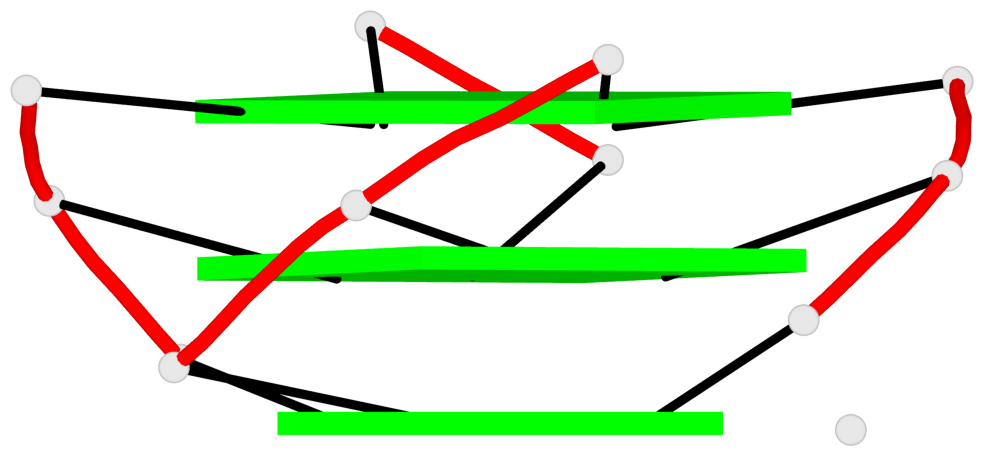
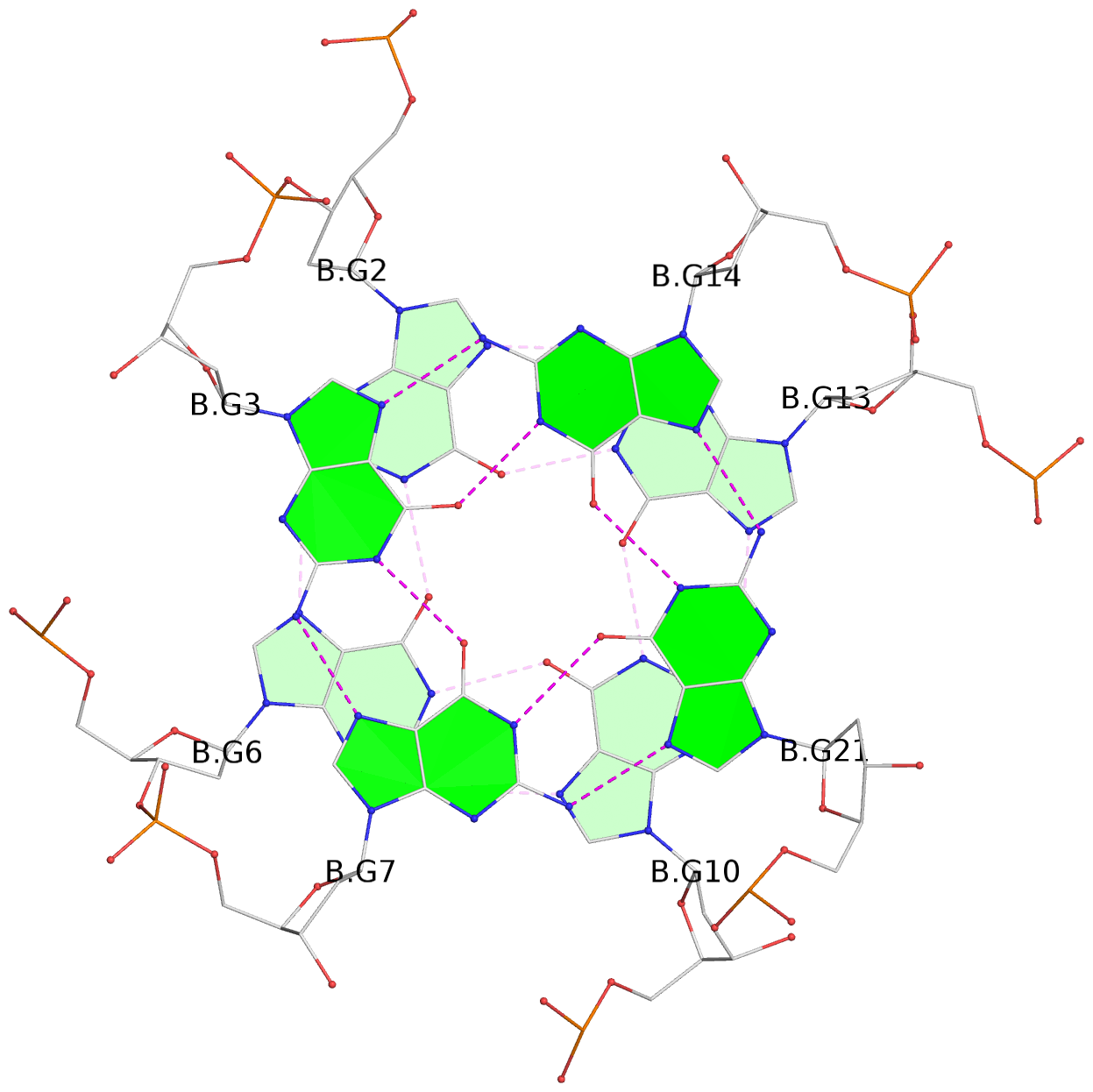
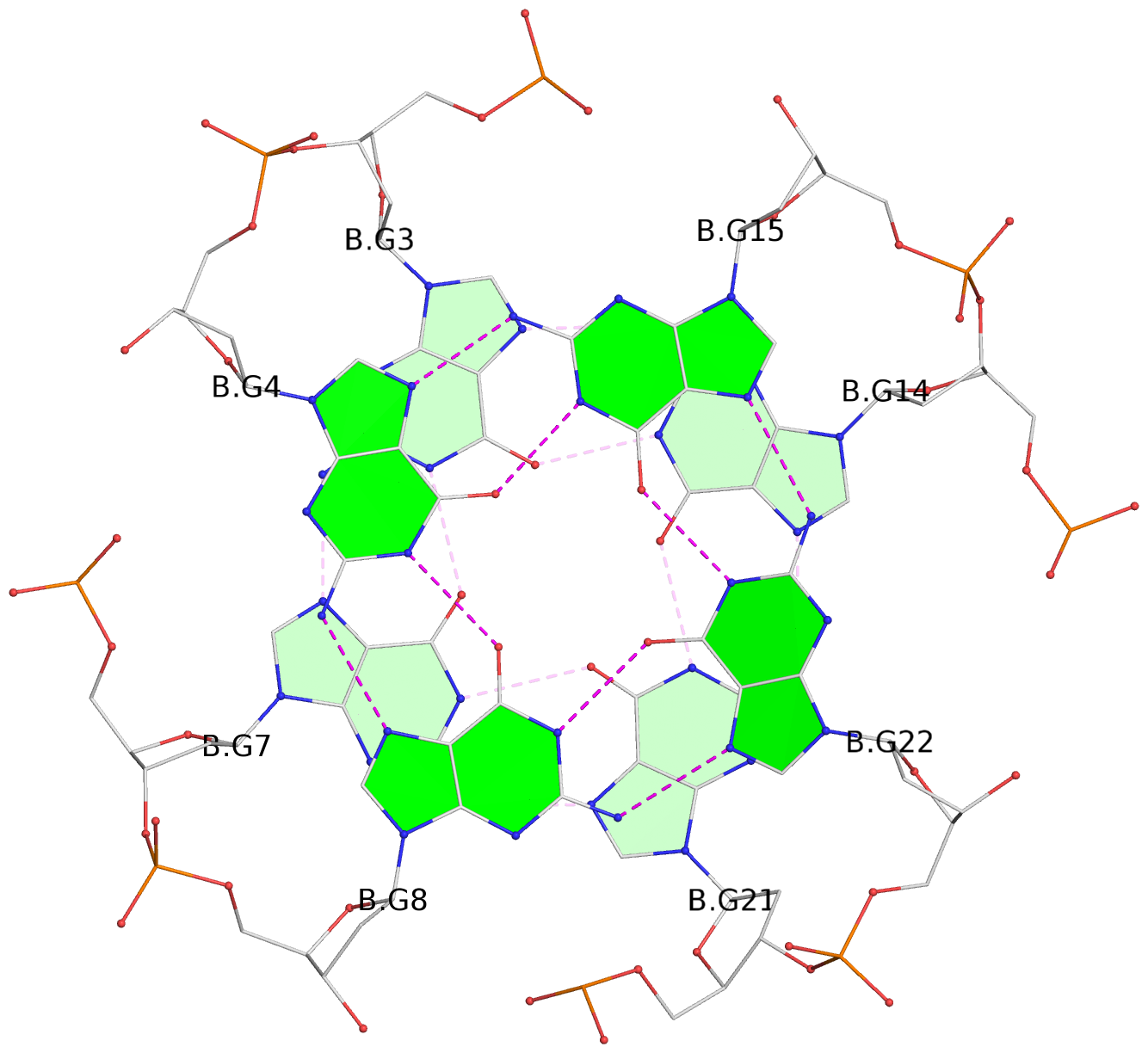
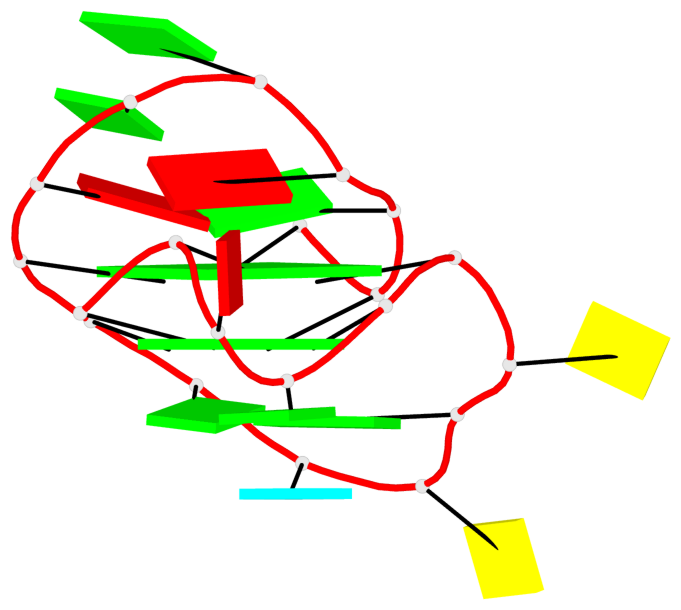
Base-block schematics in six views
List of 6 G-tetrads
1 glyco-bond=---- sugar=-3-- groove=---- planarity=0.230 type=other nts=4 GGGG A.DG2,A.DG6,A.DG10,A.DG13 2 glyco-bond=---- sugar=--3- groove=---- planarity=0.272 type=other nts=4 GGGG A.DG3,A.DG7,A.DG21,A.DG14 3 glyco-bond=---- sugar=---- groove=---- planarity=0.393 type=bowl nts=4 GGGG A.DG4,A.DG8,A.DG22,A.DG15 4 glyco-bond=---- sugar=33-. groove=---- planarity=0.229 type=other nts=4 GGGG B.DG2,B.DG6,B.DG10,B.DG13 5 glyco-bond=---- sugar=--3- groove=---- planarity=0.253 type=other nts=4 GGGG B.DG3,B.DG7,B.DG21,B.DG14 6 glyco-bond=---- sugar=---- groove=---- planarity=0.346 type=bowl nts=4 GGGG B.DG4,B.DG8,B.DG22,B.DG15
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
Helix#2, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-PX+P), parallel(4+0)
Stem#2, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-PX+P), parallel(4+0)
List of 4 non-stem G4-loops (including the two closing Gs)
1* type=V-shaped helix=#1 nts=5 GGGCG A.DG6,A.DG7,A.DG8,A.DC9,A.DG10 2 type=lateral helix=#1 nts=4 GCuG A.DG10,A.DC11,A.BRU12,A.DG13 3* type=V-shaped helix=#2 nts=5 GGGCG B.DG6,B.DG7,B.DG8,B.DC9,B.DG10 4 type=lateral helix=#2 nts=4 GCuG B.DG10,B.DC11,B.BRU12,B.DG13