Detailed DSSR results for the G-quadruplex: PDB entry 3tvb
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 3tvb
- Class
- DNA-antibiotic
- Method
- X-ray (1.08 Å)
- Summary
- A highly symmetric DNA g-4 quadruplex-drug complex
- Reference
- Clark GR, Pytel PD, Squire CJ (2012): "The high-resolution crystal structure of a parallel intermolecular DNA G-4 quadruplex/drug complex employing syn glycosyl linkages." Nucleic Acids Res., 40, 5731-5738. doi: 10.1093/nar/gks193.
- Abstract
- We have determined the X-ray structure of the complex between the DNA quadruplex d(5'-GGGG-3')(4) and daunomycin, as a potential model for studying drug-telomere interactions. The structure was solved at 1.08 Å by direct methods in space group I4. The asymmetric unit comprises a linear arrangement of one d(GGGG) strand, four daunomycin molecules, a second d(GGGG) strand facing in the opposite direction to the first, and Na and Mg cations. The crystallographic 4-fold axis generates the biological unit, which is a 12-layered structure comprising two sets of four guanine layers, with four layers each of four daunomycins stacked between the 5' faces of the two quadruplexes. The daunomycin layers fall into two groups which are novel in their mode of self assembly. The only contacts between daunomycin molecules within any one of these layers are van der Waals interactions, however there is substantial π-π stacking between successive daunomycin layers and also with adjacent guanine layers. The structure differs significantly from all other parallel d(TGGGGT)(4) quadruplexes in that the 5' guanine adopts the unusual syn glycosyl linkage, refuting the widespread belief that such conformations should all be anti. In contrast to the related d(TGGGGT)/daunomycin complex, there are no ligand-quadruplex groove insertion interactions.
- G4 notes
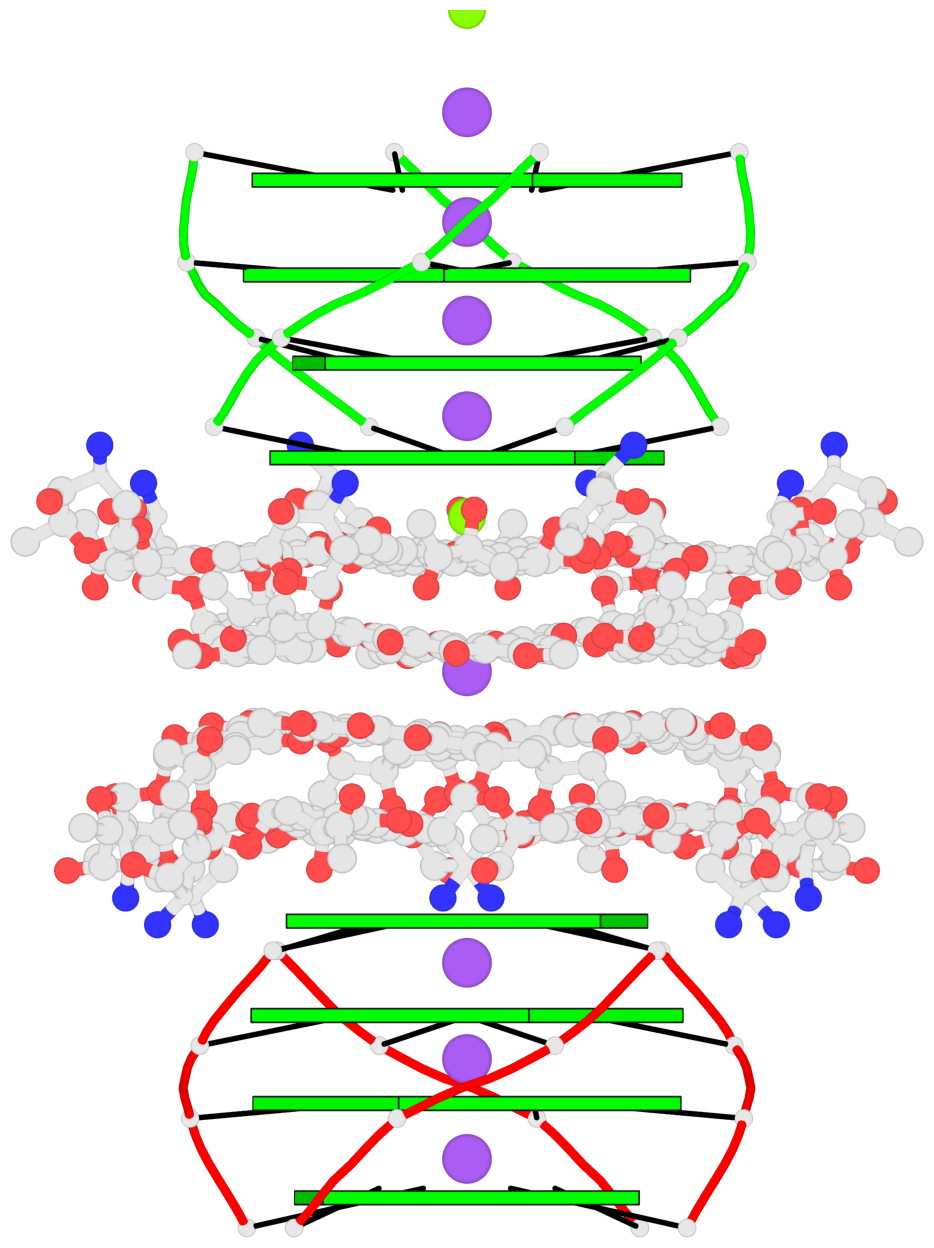
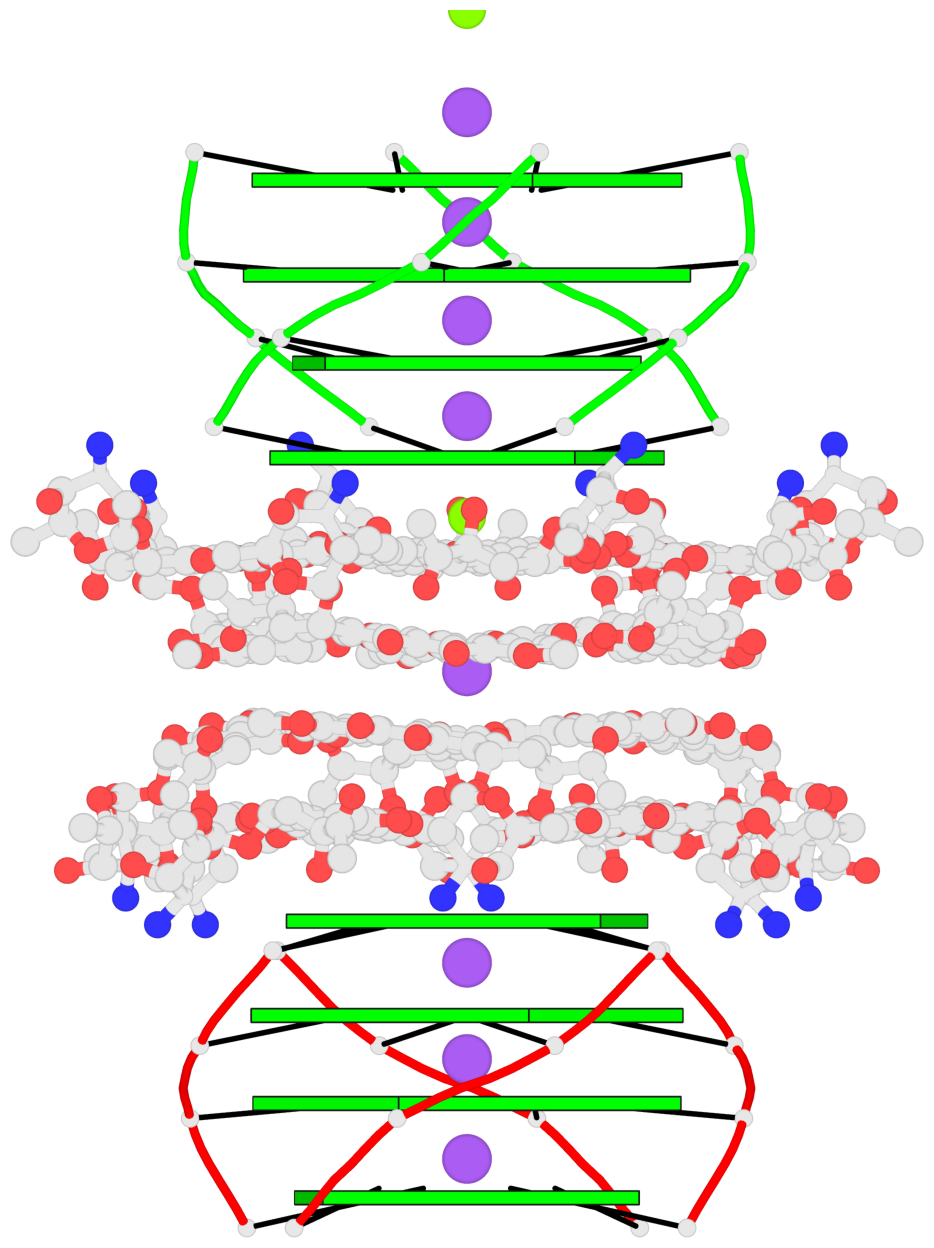
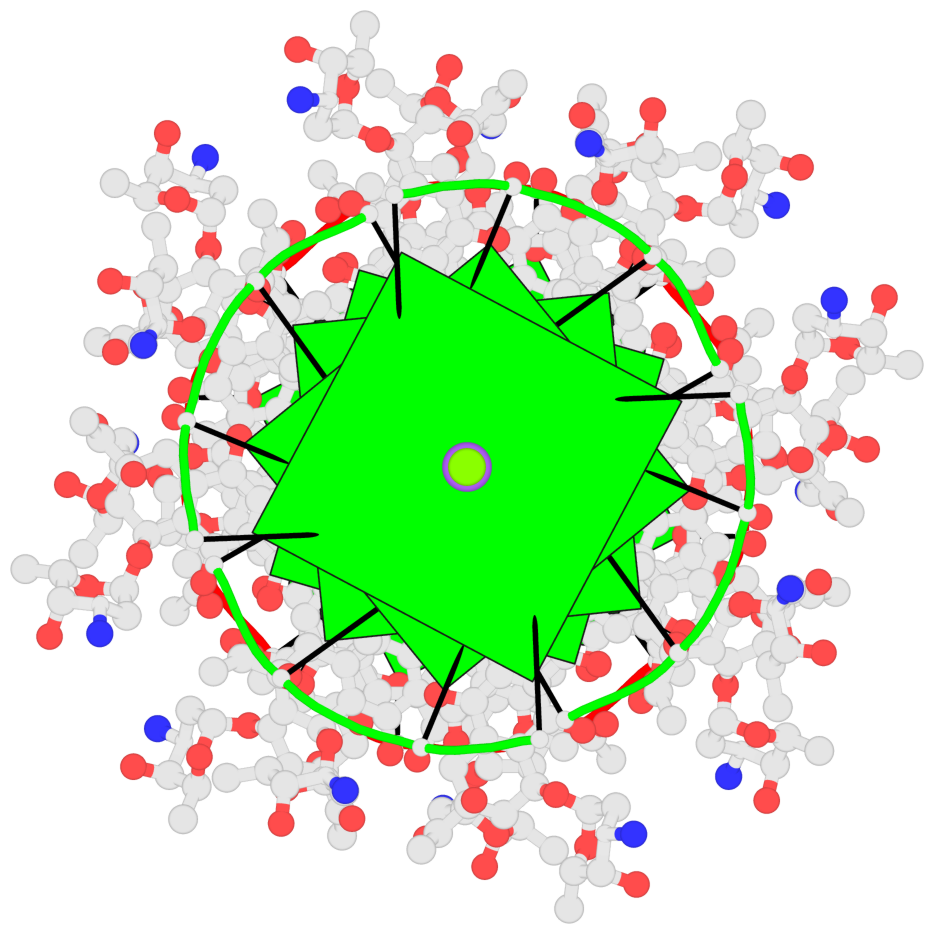
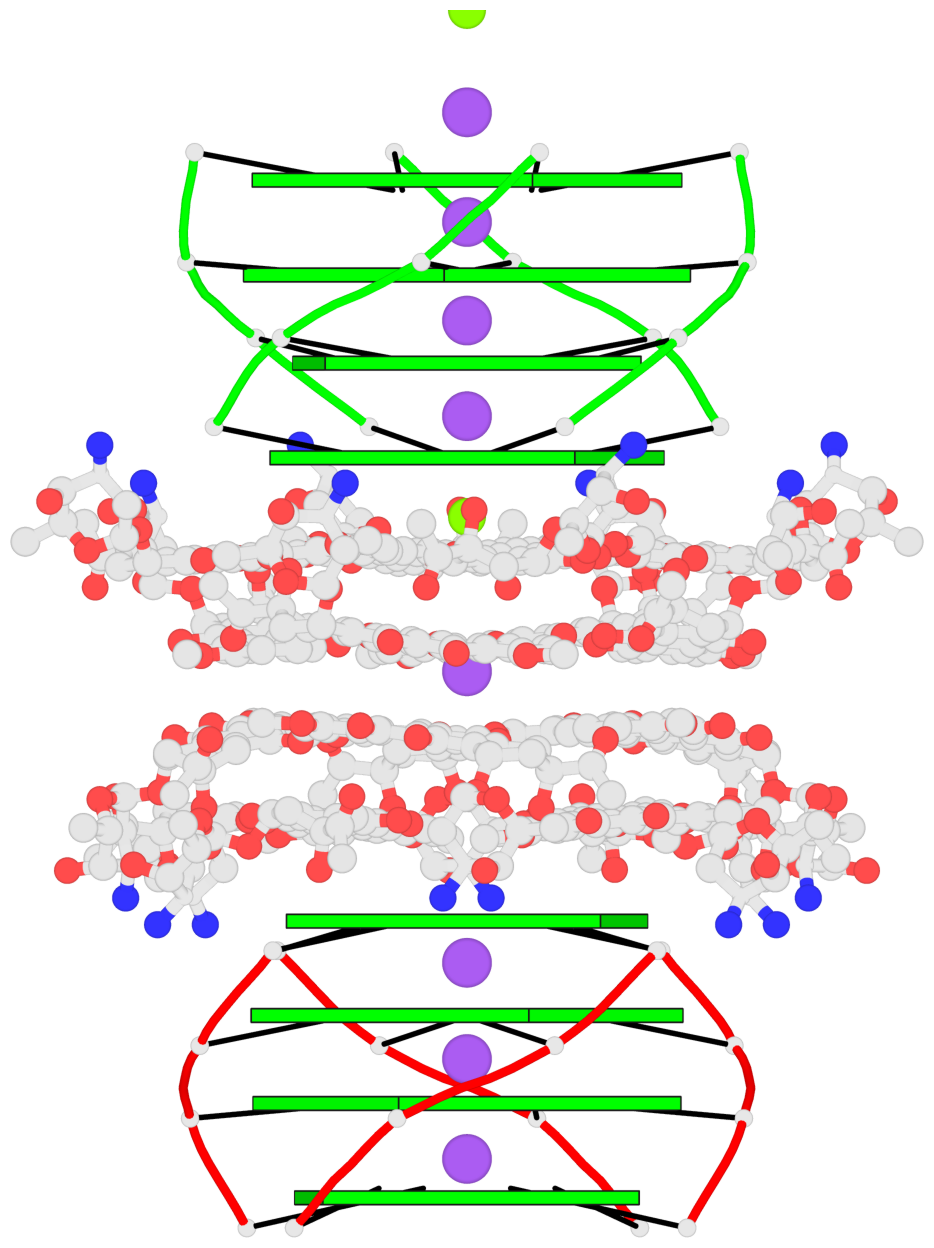
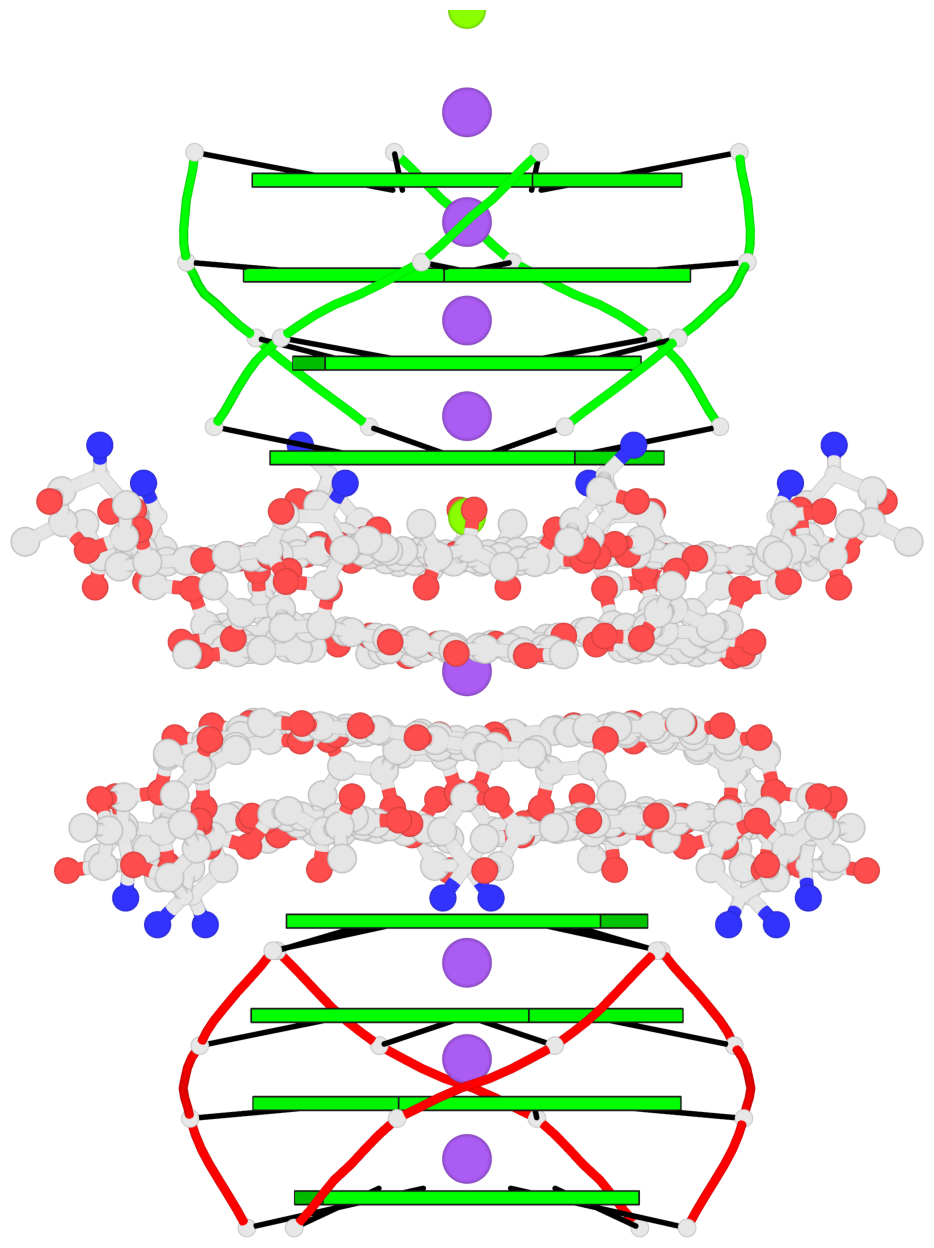
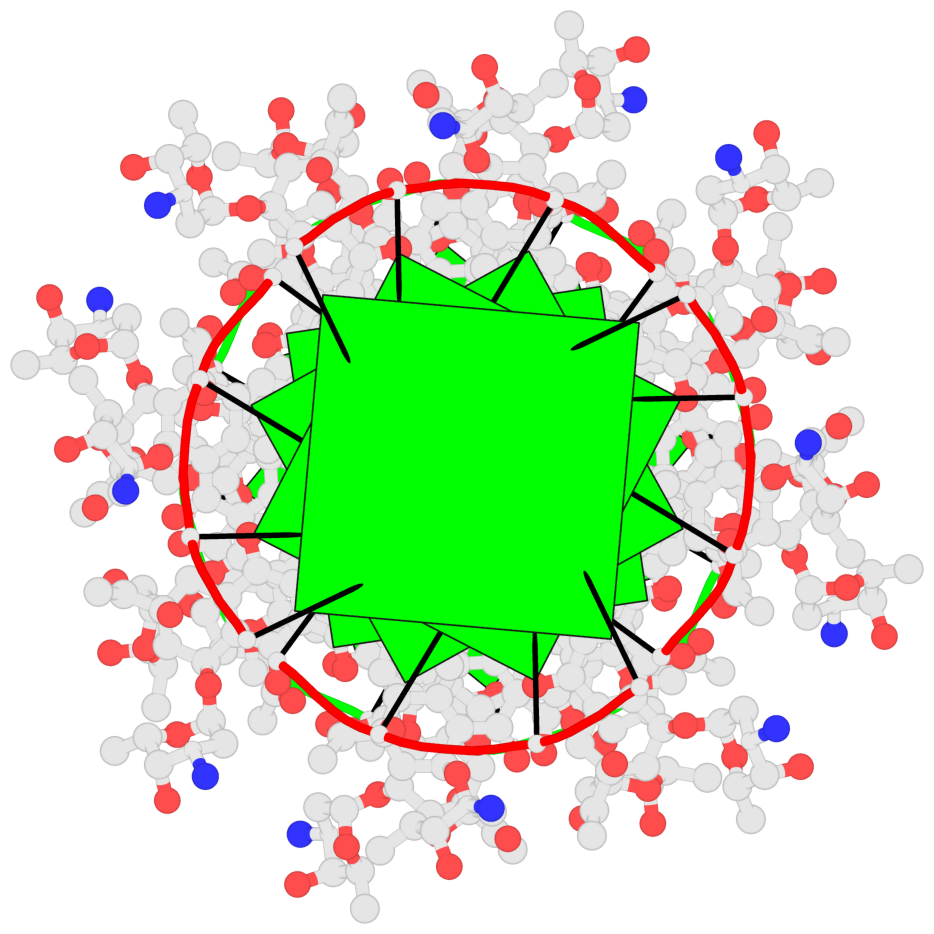
- 8 G-tetrads, 2 G4 helices, 2 G4 stems, parallel(4+0), UUUU
Base-block schematics in six views
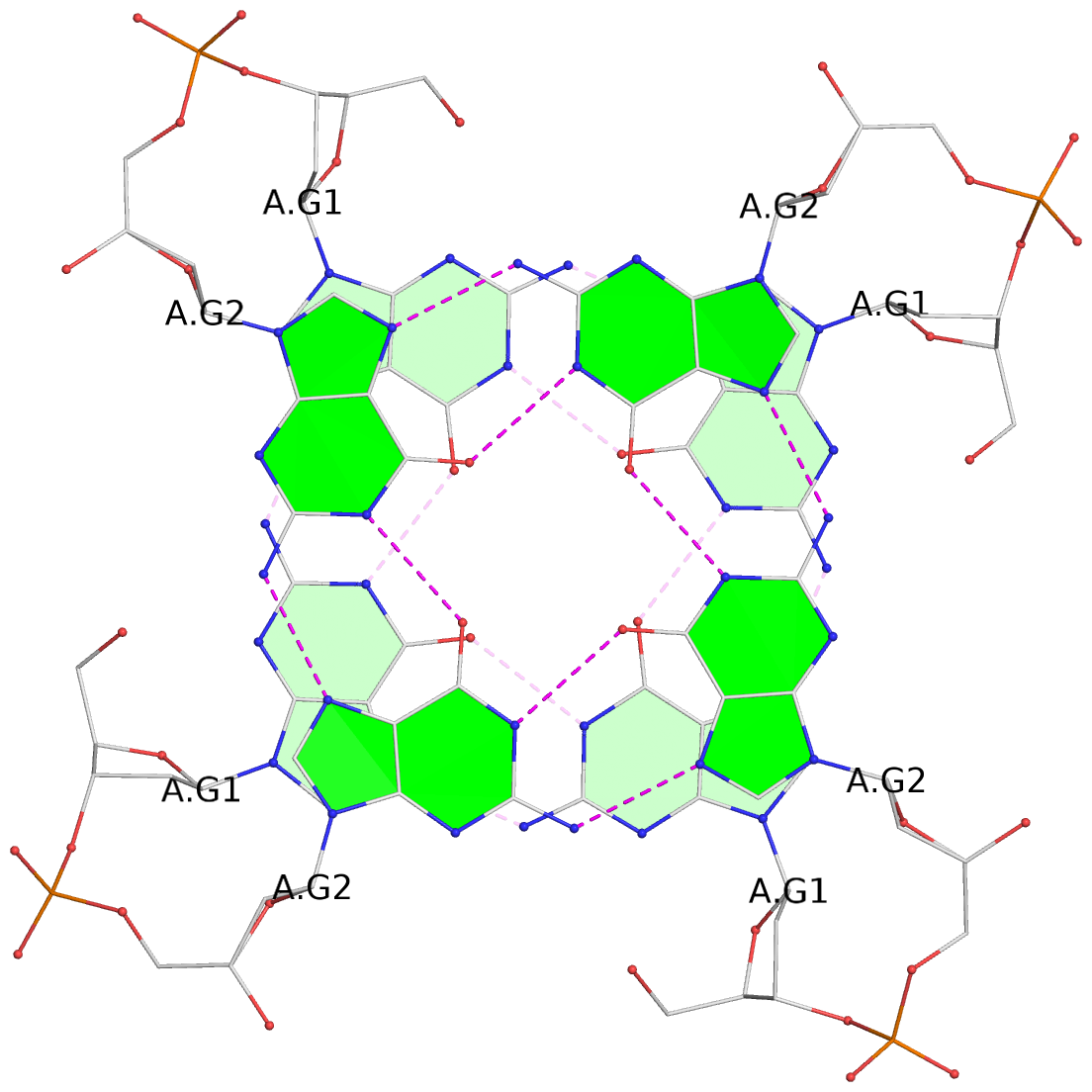
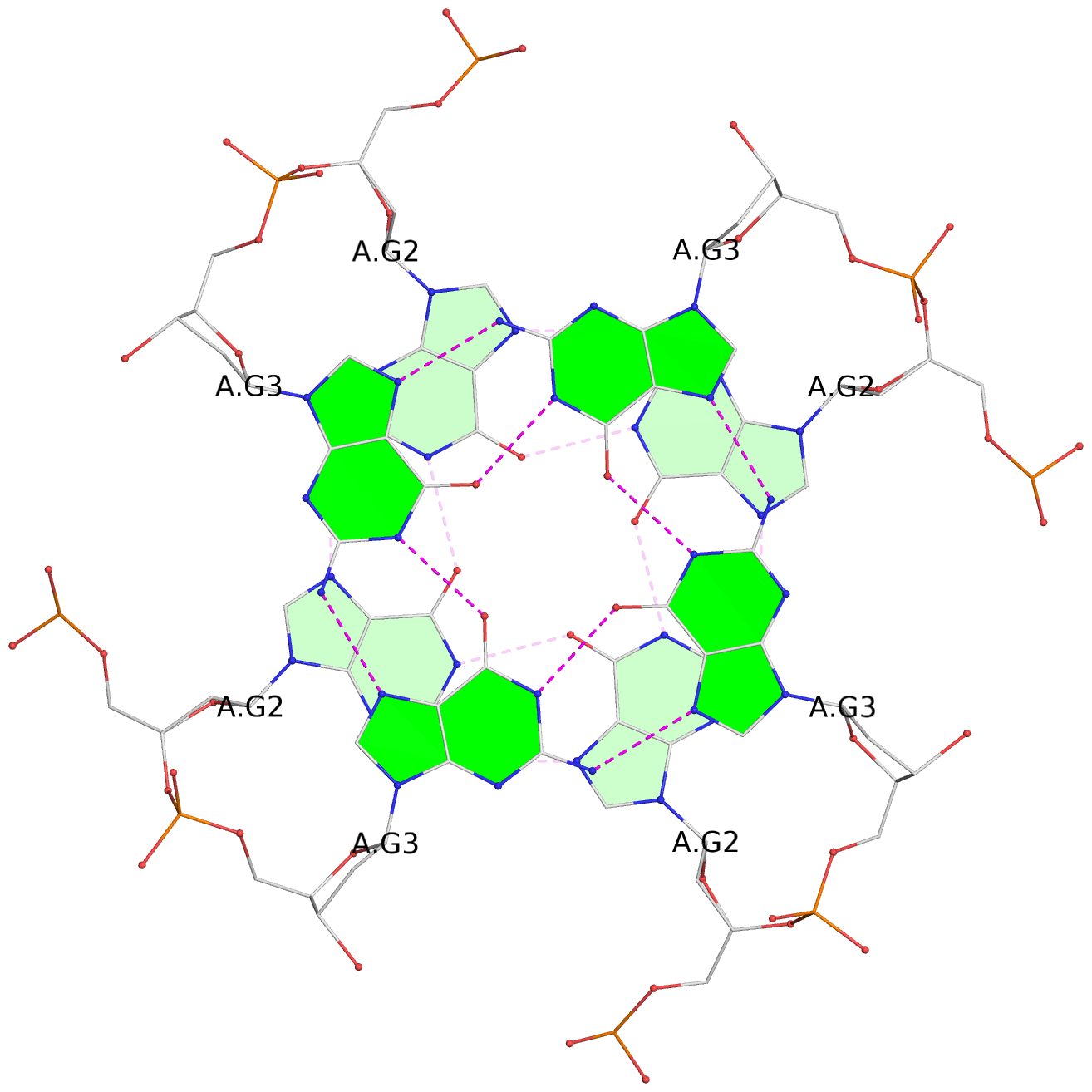
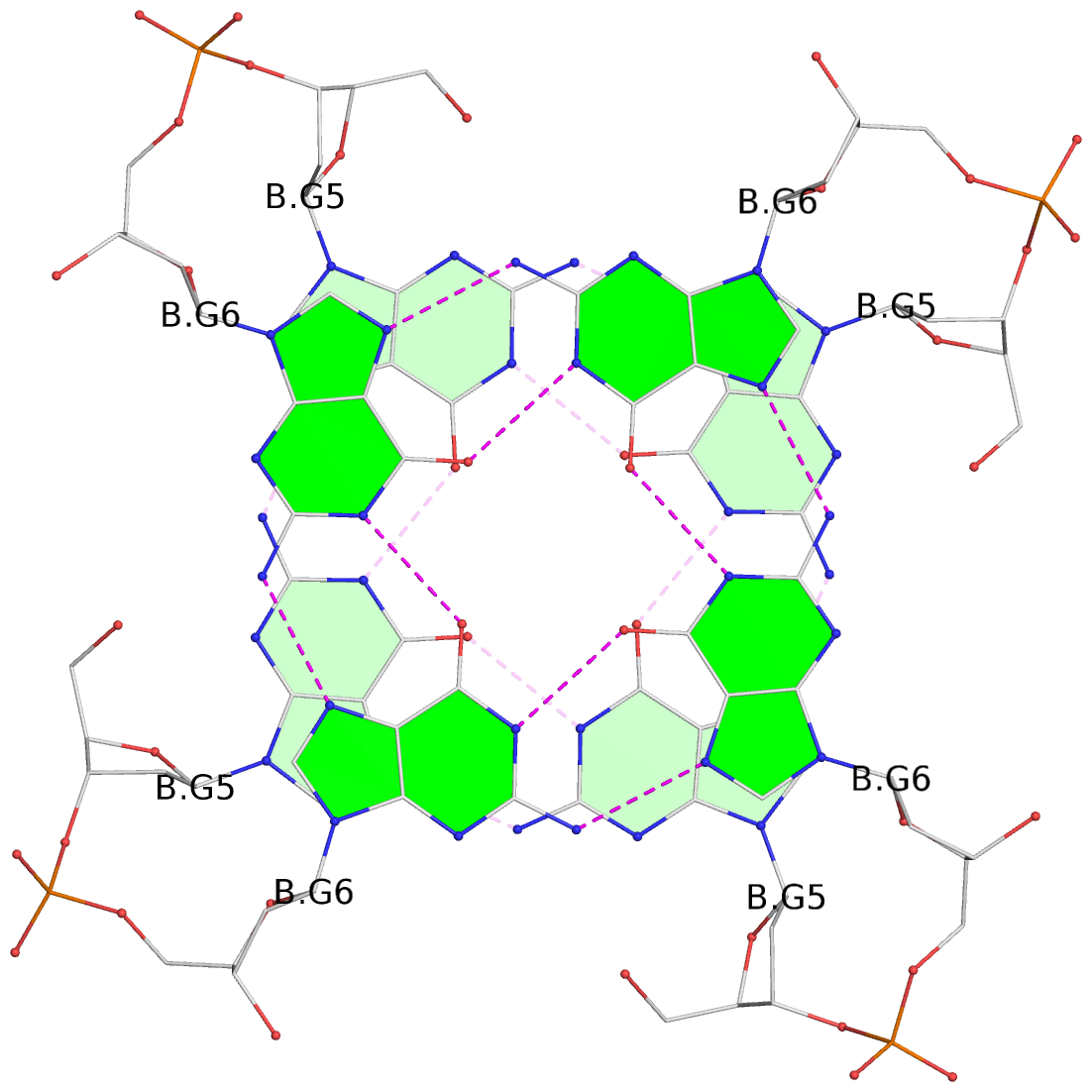
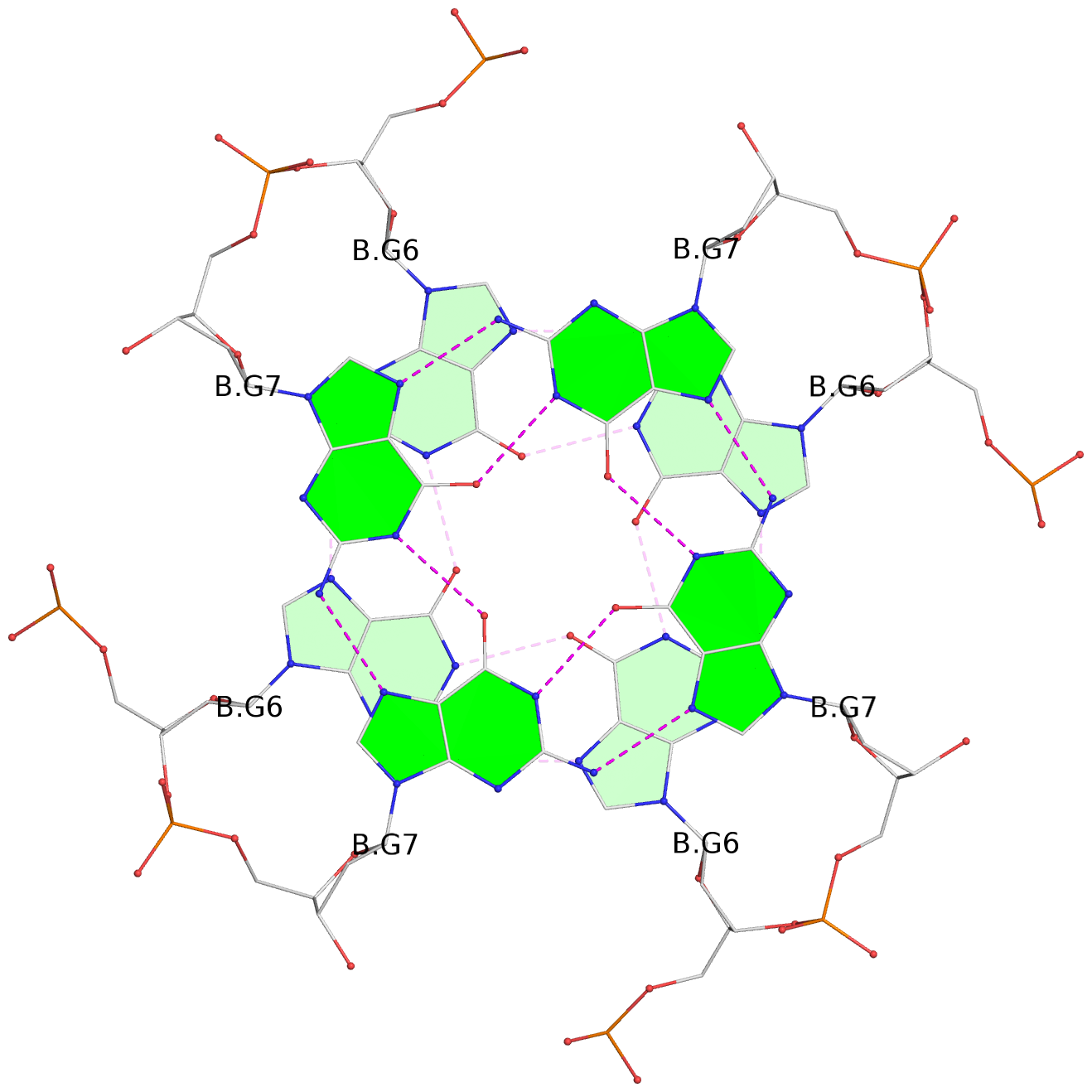
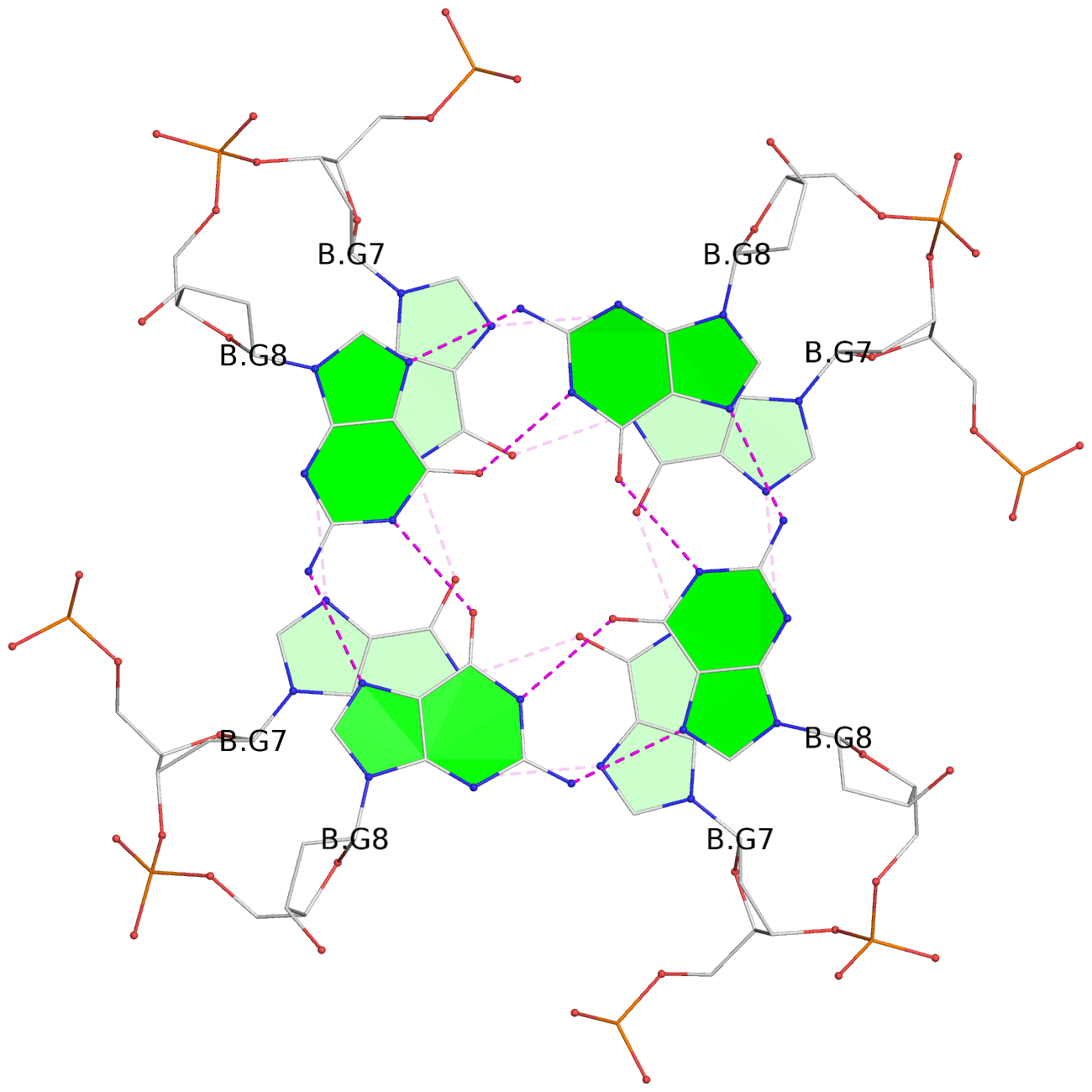
List of 8 G-tetrads
1 glyco-bond=ssss sugar=---- groove=---- planarity=0.075 type=planar nts=4 GGGG 1:A.DG1,4:A.DG1,2:A.DG1,3:A.DG1 2 glyco-bond=---- sugar=---- groove=---- planarity=0.198 type=other nts=4 GGGG 1:A.DG2,4:A.DG2,2:A.DG2,3:A.DG2 3 glyco-bond=---- sugar=---- groove=---- planarity=0.108 type=planar nts=4 GGGG 1:A.DG3,4:A.DG3,2:A.DG3,3:A.DG3 4 glyco-bond=---- sugar=---- groove=---- planarity=0.312 type=bowl nts=4 GGGG 1:A.DG4,4:A.DG4,2:A.DG4,3:A.DG4 5 glyco-bond=ssss sugar=---- groove=---- planarity=0.060 type=planar nts=4 GGGG 1:B.DG5,3:B.DG5,2:B.DG5,4:B.DG5 6 glyco-bond=---- sugar=---- groove=---- planarity=0.163 type=other nts=4 GGGG 1:B.DG6,3:B.DG6,2:B.DG6,4:B.DG6 7 glyco-bond=---- sugar=---- groove=---- planarity=0.098 type=planar nts=4 GGGG 1:B.DG7,3:B.DG7,2:B.DG7,4:B.DG7 8 glyco-bond=---- sugar=---- groove=---- planarity=0.311 type=bowl nts=4 GGGG 1:B.DG8,3:B.DG8,2:B.DG8,4:B.DG8
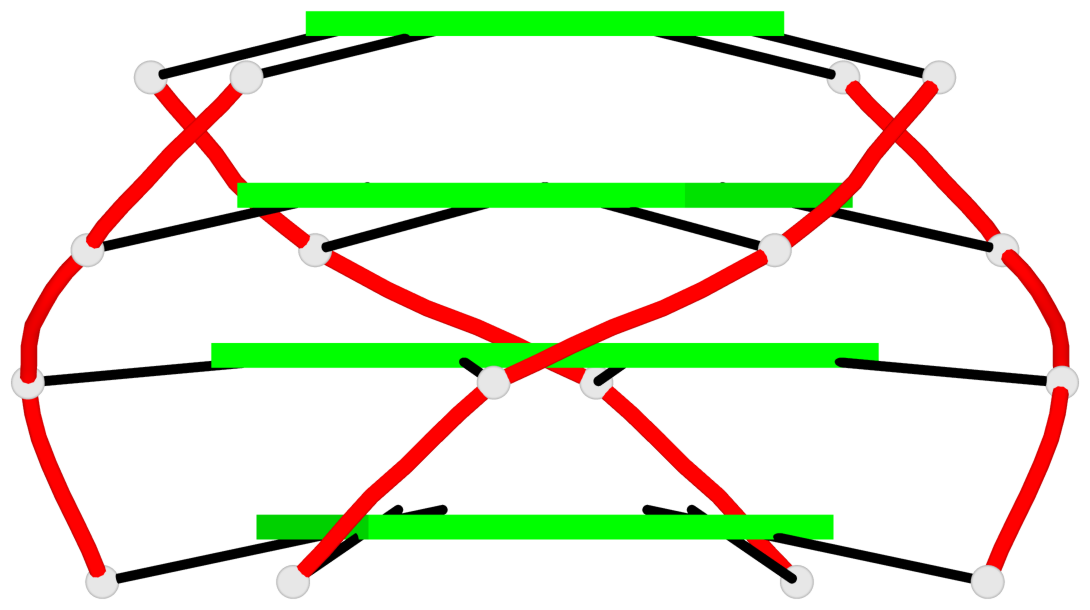
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 1 stem
Helix#2, 4 G-tetrad layers, inter-molecular, with 1 stem
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.