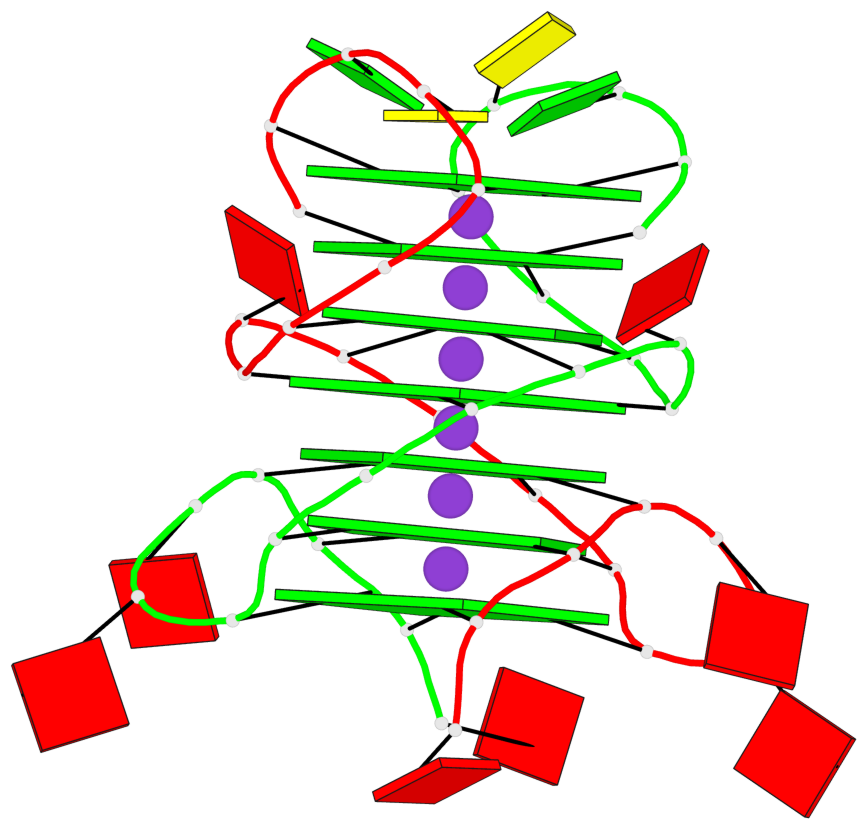
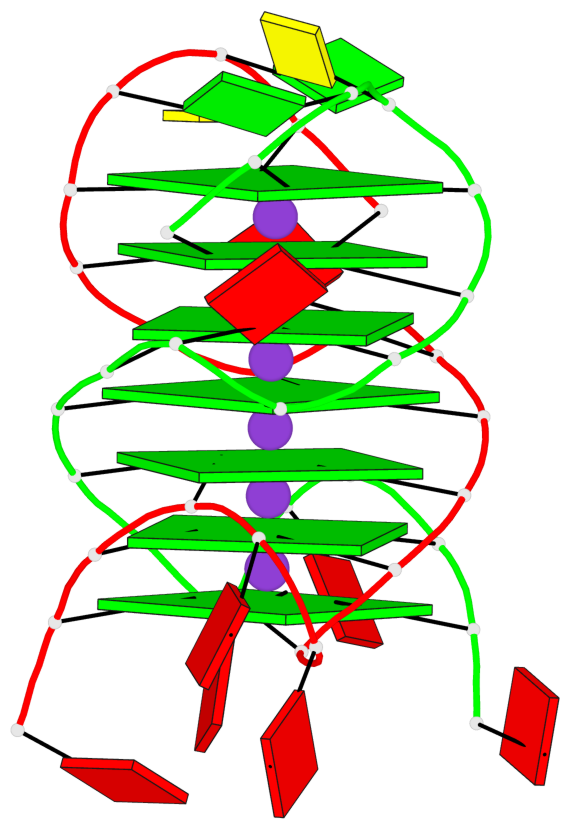
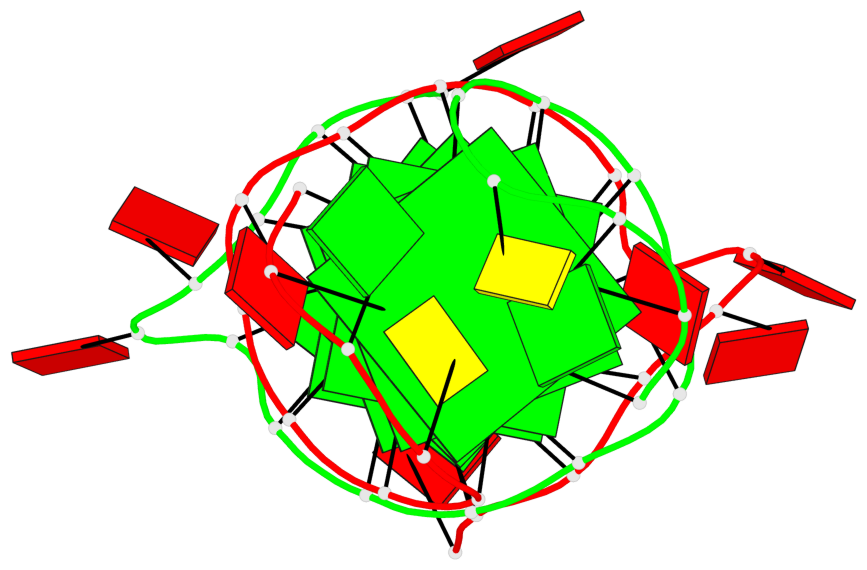
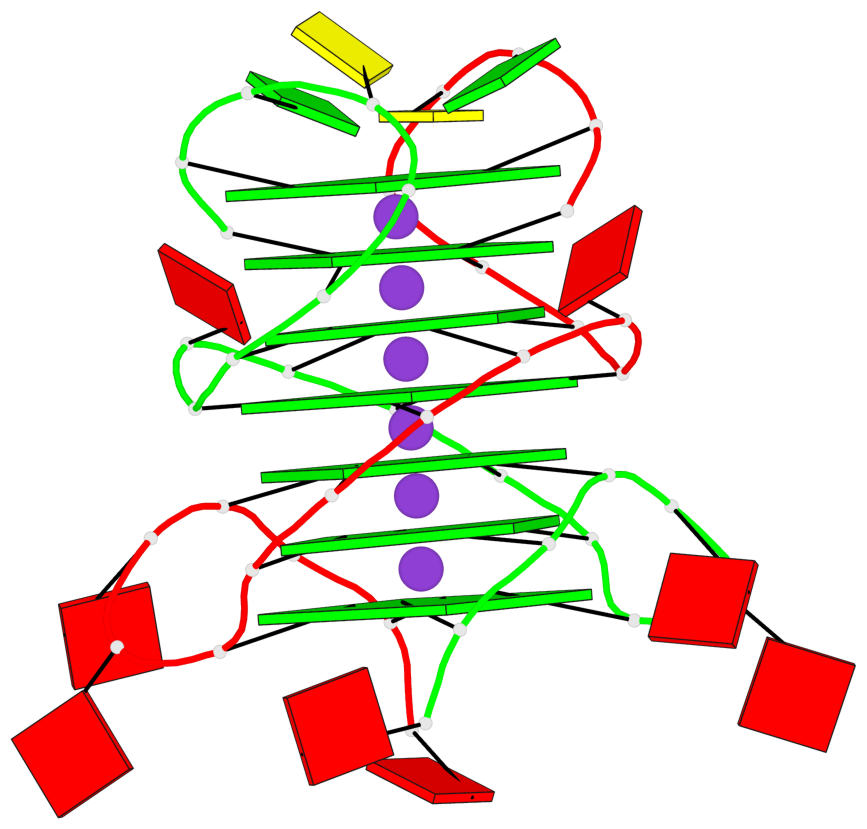
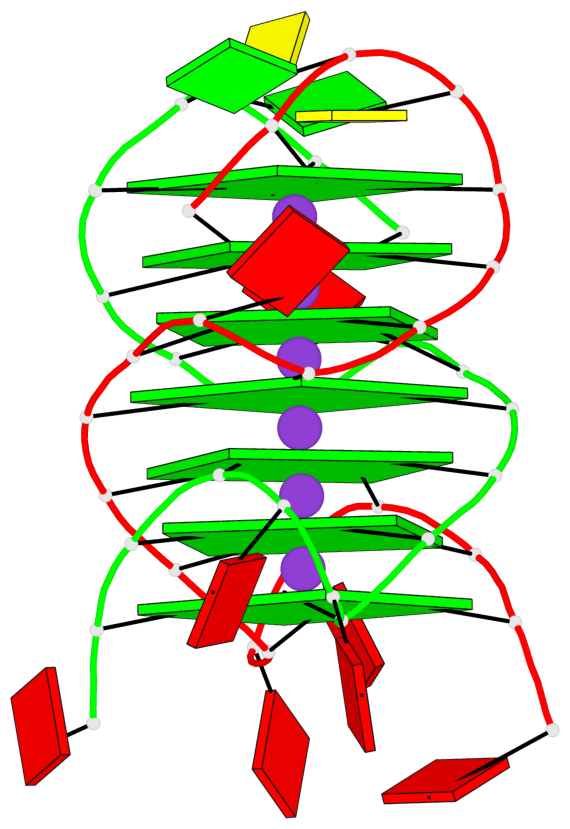
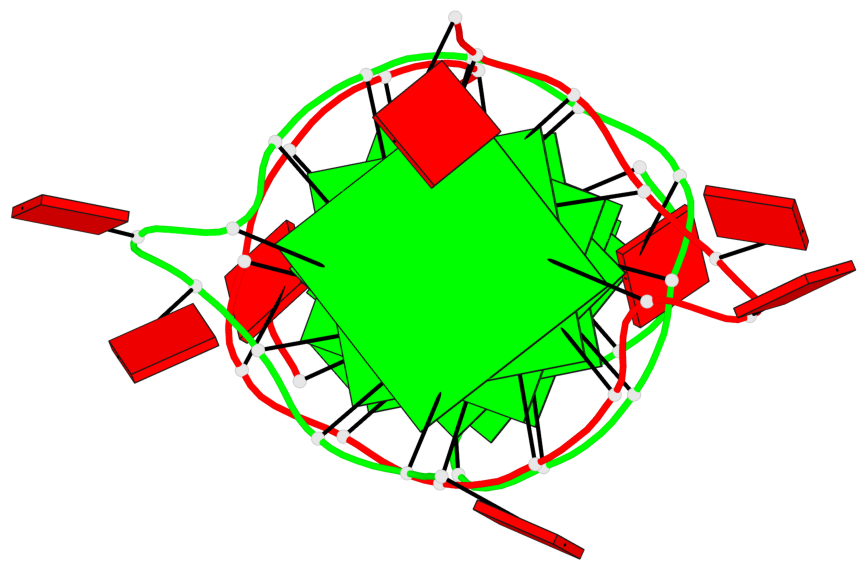
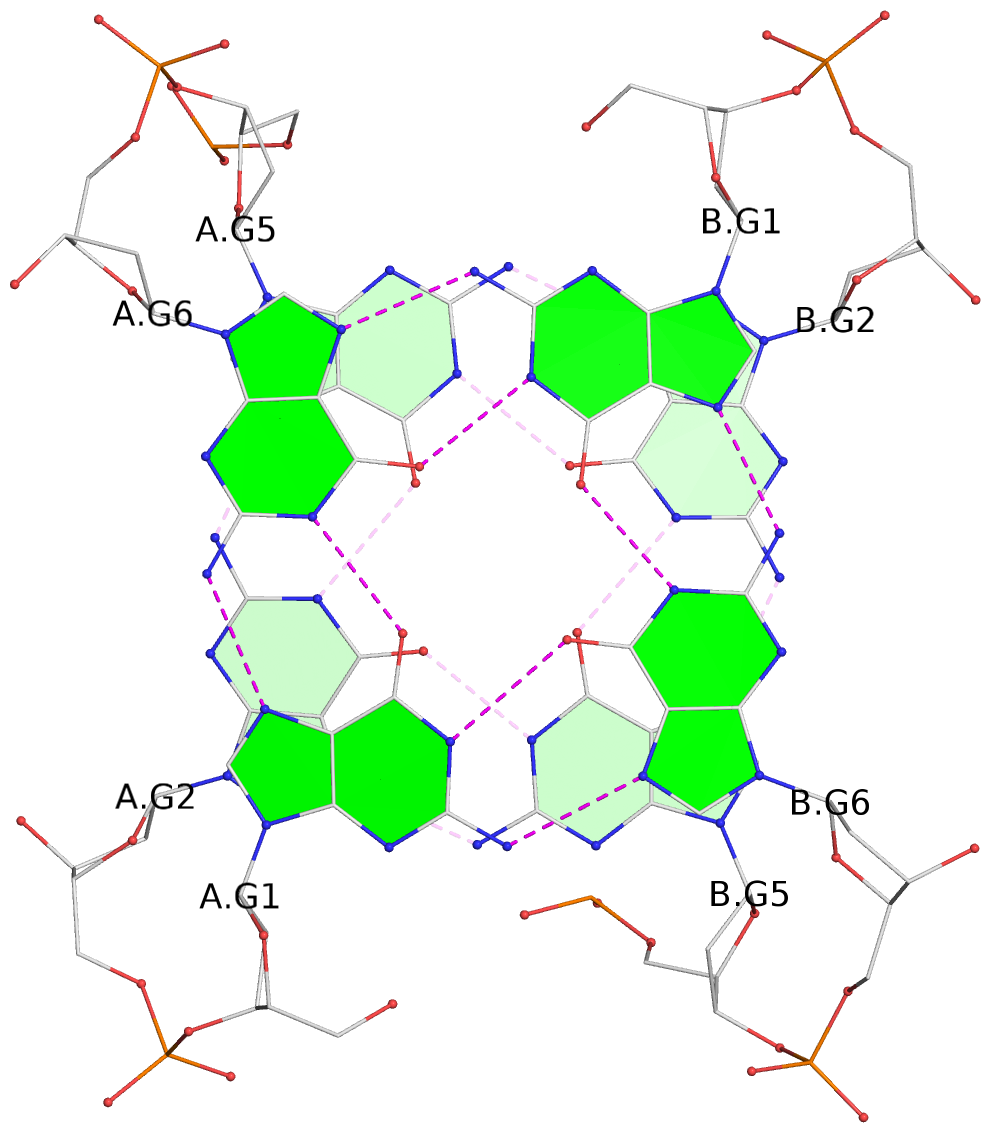
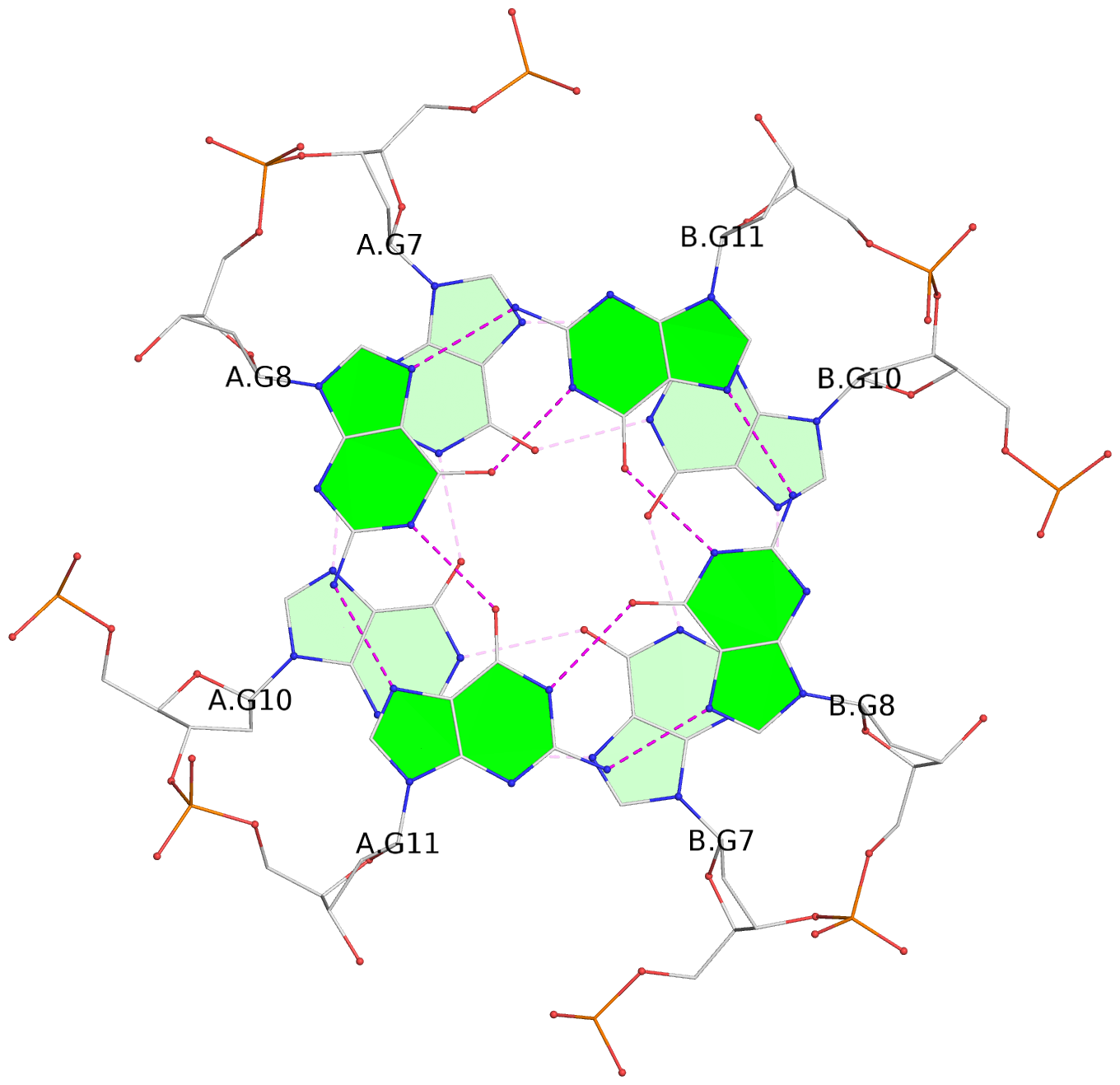
Detailed DSSR results for the G-quadruplex: PDB entry 4h29
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 4h29
- Class
- DNA
- Method
- X-ray (1.991 Å)
- Summary
- B-raf dimer DNA quadruplex
- Reference
- Wei D, Todd AK, Zloh M, Gunaratnam M, Parkinson GN, Neidle S (2013): "Crystal Structure of a Promoter Sequence in the B-raf Gene Reveals an Intertwined Dimer Quadruplex." J.Am.Chem.Soc., 135, 19319-19329. doi: 10.1021/ja4101358.
- Abstract
- The sequence d(GGGCGGGGAGGGGGAAGGGA) occurs in the promoter region of the B-raf gene. An X-ray crystallographic study has found that this forms an unprecedented dimeric quadruplex arrangement, with a core of seven consecutive G-quartets and an uninterrupted run of six potassium ions in the central channel of the quadruplex. Analogy with previously reported promoter quadruplexes had initially suggested that in common with these a monomeric quadruplex was to be expected. The structure has a distorted G·C·G·C base quartet at one end and four flipped-out adenosine nucleosides at the other. The only loops in the structure are formed by the cytosine and by the three adenosines within the sequence, with all of the guanosines participating in G-quartet formation. Solution UV and circular dichroism data are in accord with a stable quadruple arrangement being formed. 1D NMR data, together with gel electrophoresis measurements, are consistent with a dimer being the dominant species in potassium solution. A single-chain intramolecular quadruplex has been straightforwardly constructed using molecular modeling, by means of a six-nucleotide sequence joining 3' and 5' ends of each strand in the dimer. A human genomic database search has revealed a number of sequences containing eight or more consecutive short G-tracts, suggesting that such intramolecular quadruplexes could be formed within the human genome.
- G4 notes
- 7 G-tetrads, 1 G4 helix, 3 G4 stems, 1 G4 coaxial stack, (2+2), UDUD; parallel(4+0), UUUU, coaxial interfaces: mixed; 3'/5'
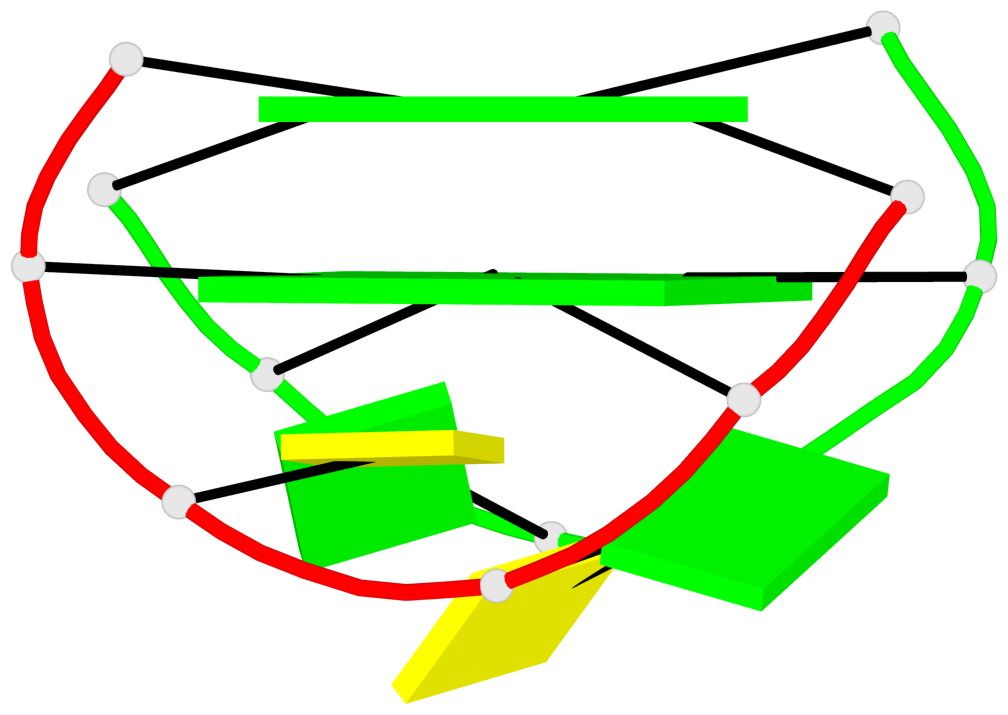
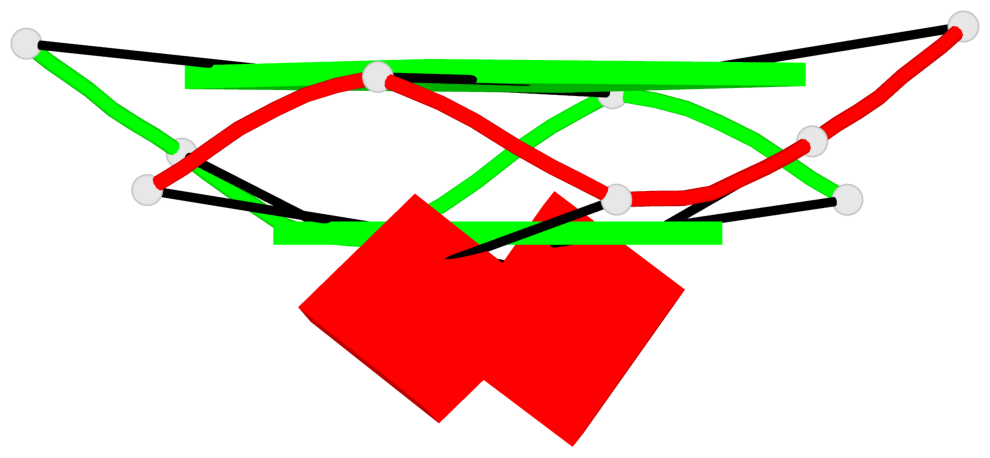
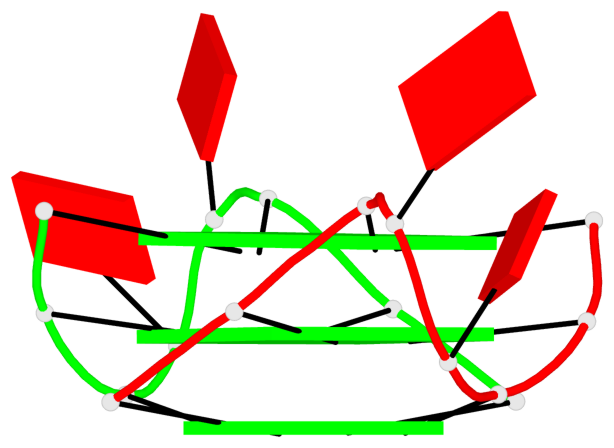
Base-block schematics in six views
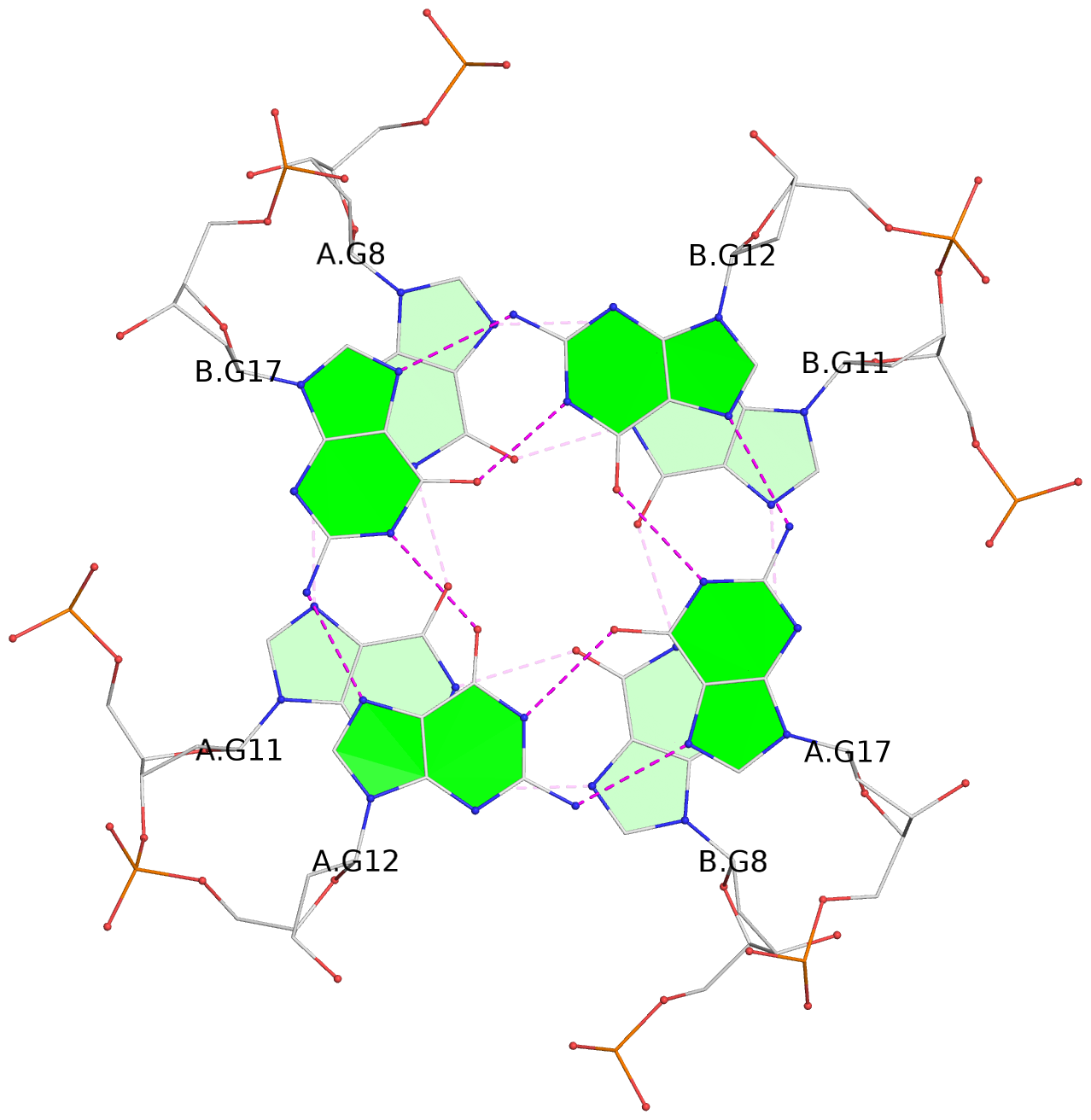
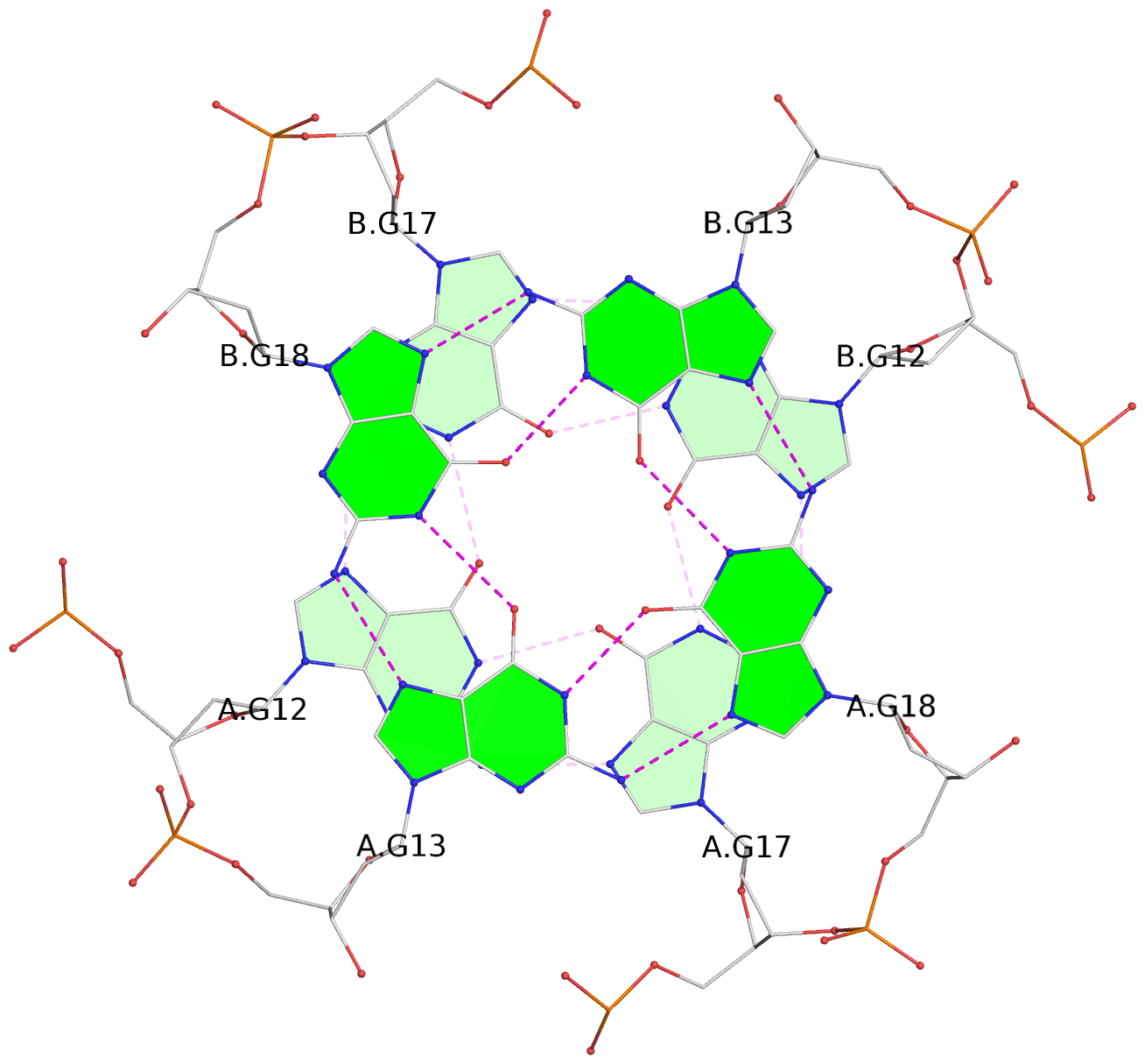
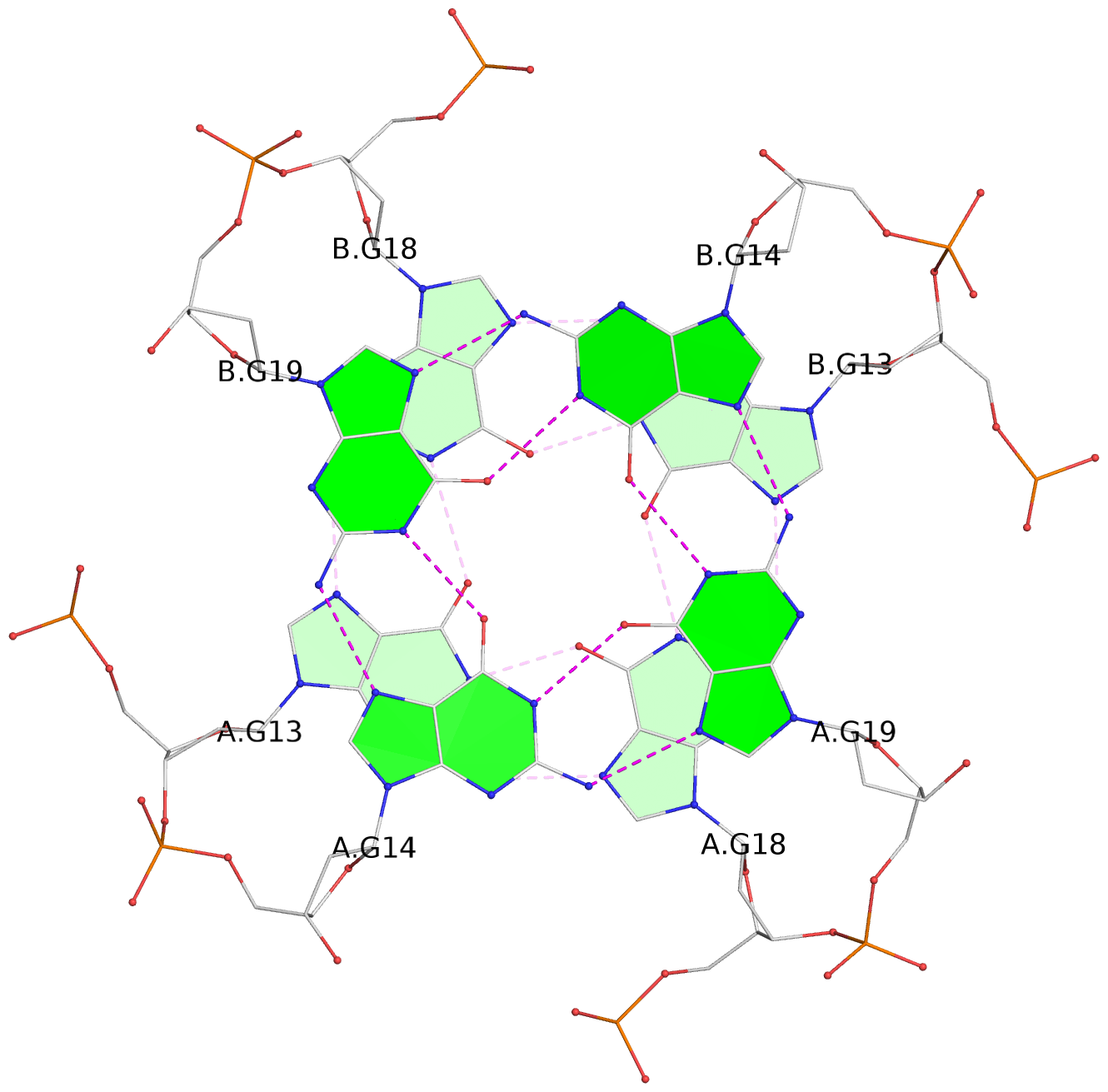
List of 7 G-tetrads
1 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.391 type=saddle nts=4 GGGG A.DG1,A.DG6,B.DG1,B.DG6 2 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.468 type=bowl-2 nts=4 GGGG A.DG2,A.DG5,B.DG2,B.DG5 3 glyco-bond=---- sugar=-.-- groove=---- planarity=0.159 type=planar nts=4 GGGG A.DG7,A.DG10,B.DG7,B.DG10 4 glyco-bond=---- sugar=---- groove=---- planarity=0.177 type=bowl-2 nts=4 GGGG A.DG8,A.DG11,B.DG8,B.DG11 5 glyco-bond=---- sugar=---- groove=---- planarity=0.187 type=bowl-2 nts=4 GGGG A.DG12,A.DG17,B.DG12,B.DG17 6 glyco-bond=---- sugar=---- groove=---- planarity=0.161 type=other nts=4 GGGG A.DG13,A.DG18,B.DG13,B.DG18 7 glyco-bond=---- sugar=---- groove=---- planarity=0.311 type=bowl nts=4 GGGG A.DG14,A.DG19,B.DG14,B.DG19
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 7 G-tetrad layers, inter-molecular, with 3 stems
List of 3 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 2 loops, inter-molecular, UDUD, anti-parallel, (2+2)
Stem#2, 2 G-tetrad layers, 2 loops, inter-molecular, UUUU, parallel, parallel(4+0)
Stem#3, 3 G-tetrad layers, 2 loops, inter-molecular, UUUU, parallel, parallel(4+0)
List of 1 G4 coaxial stack
1 G4 helix#1 contains 3 G4 stems: [#1,#2,#3] [mixed,3'/5']