Detailed DSSR results for the G-quadruplex: PDB entry 4l0a
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 4l0a
- Class
- DNA, RNA
- Method
- X-ray (1.7 Å)
- Summary
- X-ray structure of an all lna quadruplex
- Reference
- Russo Krauss I, Parkinson GN, Merlino A, Mattia CA, Randazzo A, Novellino E, Mazzarella L, Sica F (2014): "A regular thymine tetrad and a peculiar supramolecular assembly in the first crystal structure of an all-LNA G-quadruplex." Acta Crystallogr.,Sect.D, 70, 362-370. doi: 10.1107/S1399004713028095.
- Abstract
- Locked nucleic acids (LNAs) are formed by bicyclic ribonucleotides where the O2' and C4' atoms are linked through a methylene bridge and the sugar is blocked in a 3'-endo conformation. They represent a promising tool for therapeutic and diagnostic applications and are characterized by higher thermal stability and nuclease resistance with respect to their natural counterparts. However, structural descriptions of LNA-containing quadruplexes are rather limited, since few NMR models have been reported in the literature. Here, the first crystallographically derived model of an all-LNA-substituted quadruplex-forming sequence 5'-TGGGT-3' is presented refined at 1.7 Å resolution. This high-resolution crystallographic analysis reveals a regular parallel G-quadruplex arrangement terminating in a well defined thymine tetrad at the 3'-end. The detailed picture of the hydration pattern reveals LNA-specific features in the solvent distribution. Interestingly, two closely packed quadruplexes are present in the asymmetric unit. They face one another with their 3'-ends giving rise to a compact higher-order structure. This new assembly suggests a possible way in which sequential quadruplexes can be disposed in the crowded cell environment. Furthermore, as the formation of ordered structures by molecular self-assembly is an effective strategy to obtain nanostructures, this study could open the way to the design of a new class of LNA-based building blocks for nanotechnology.
- G4 notes
- 6 G-tetrads, 2 G4 helices, 2 G4 stems, parallel(4+0), UUUU
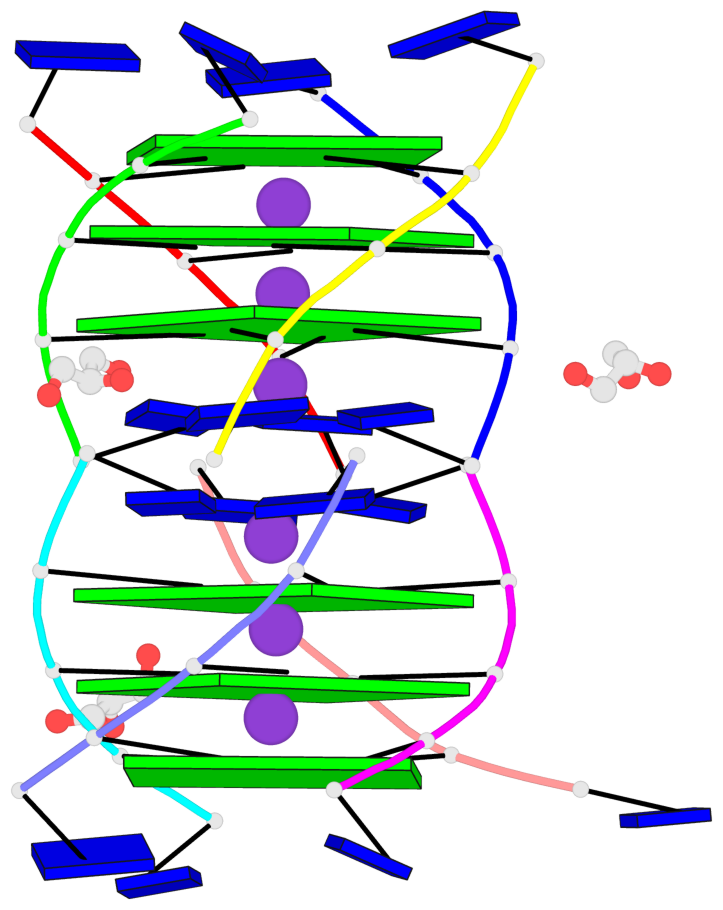
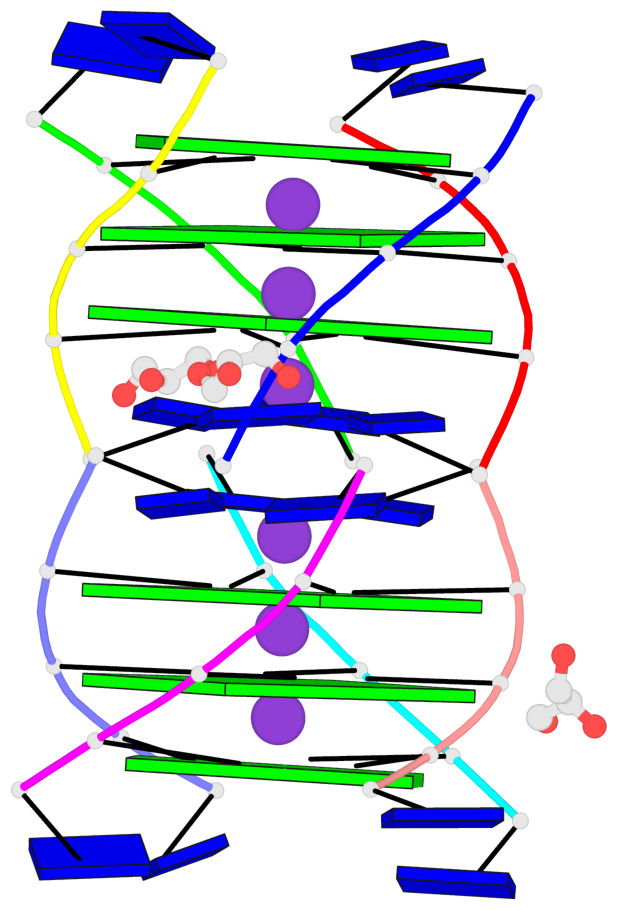
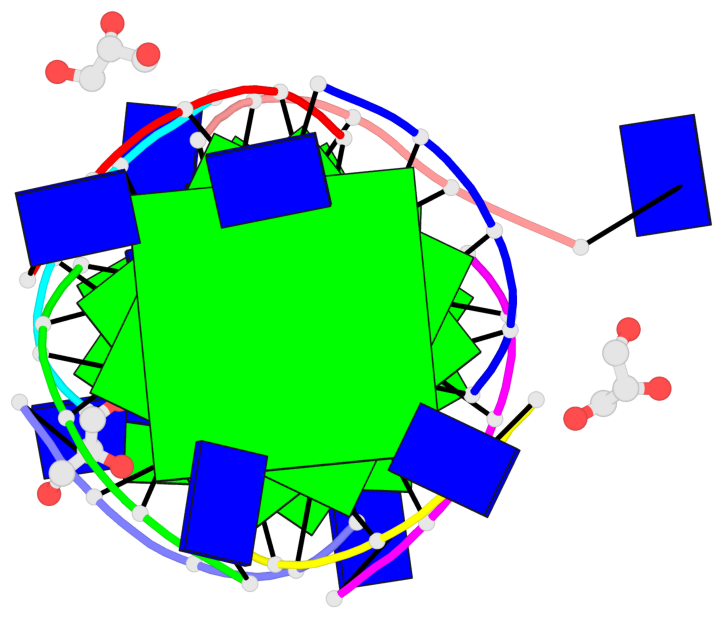
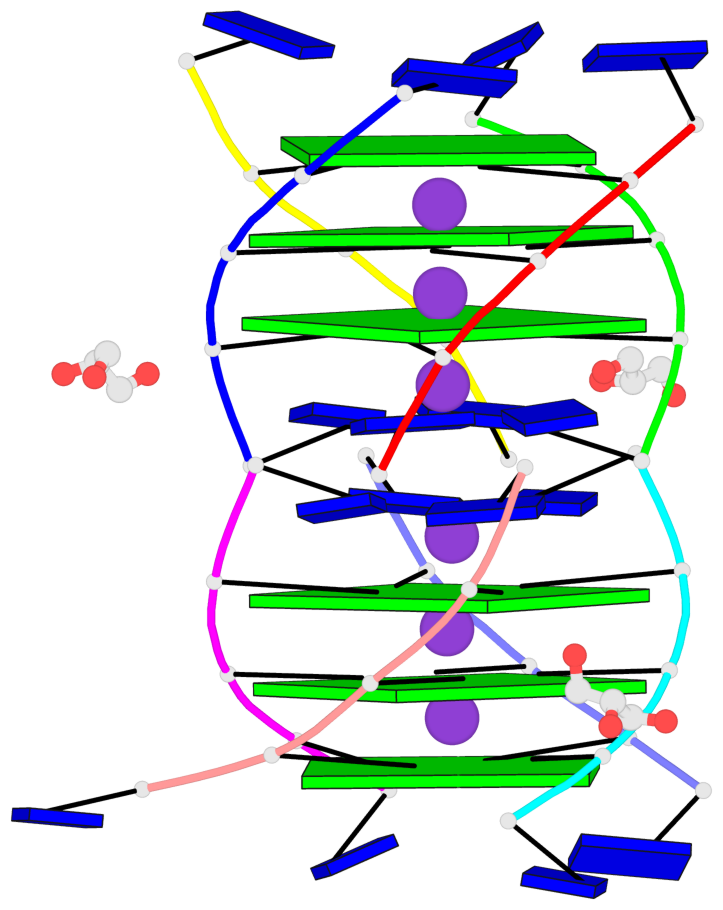
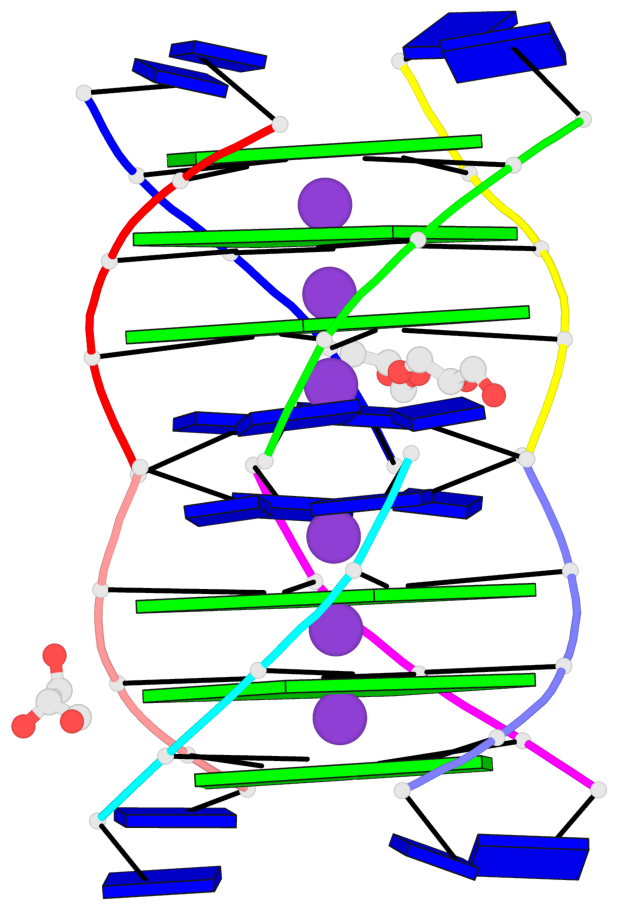
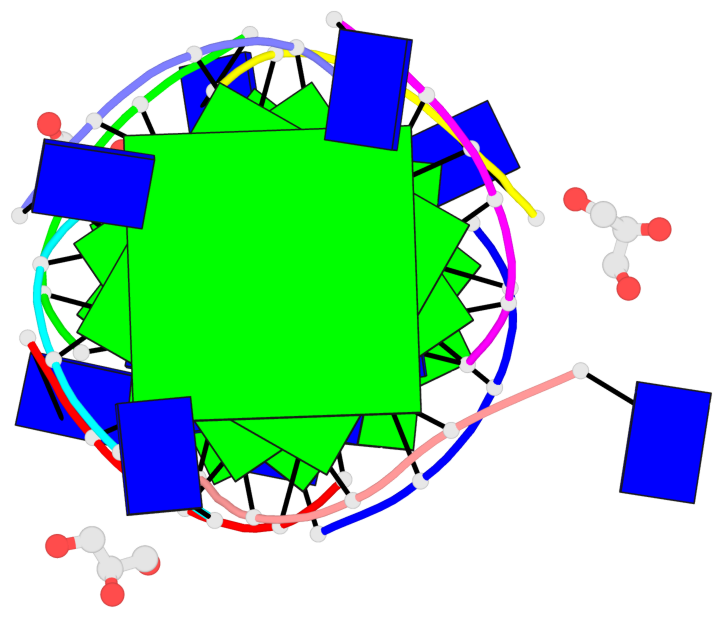
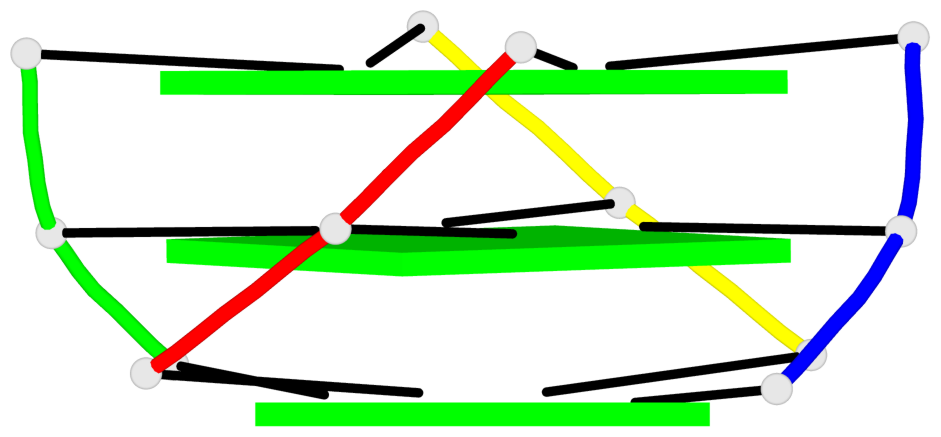
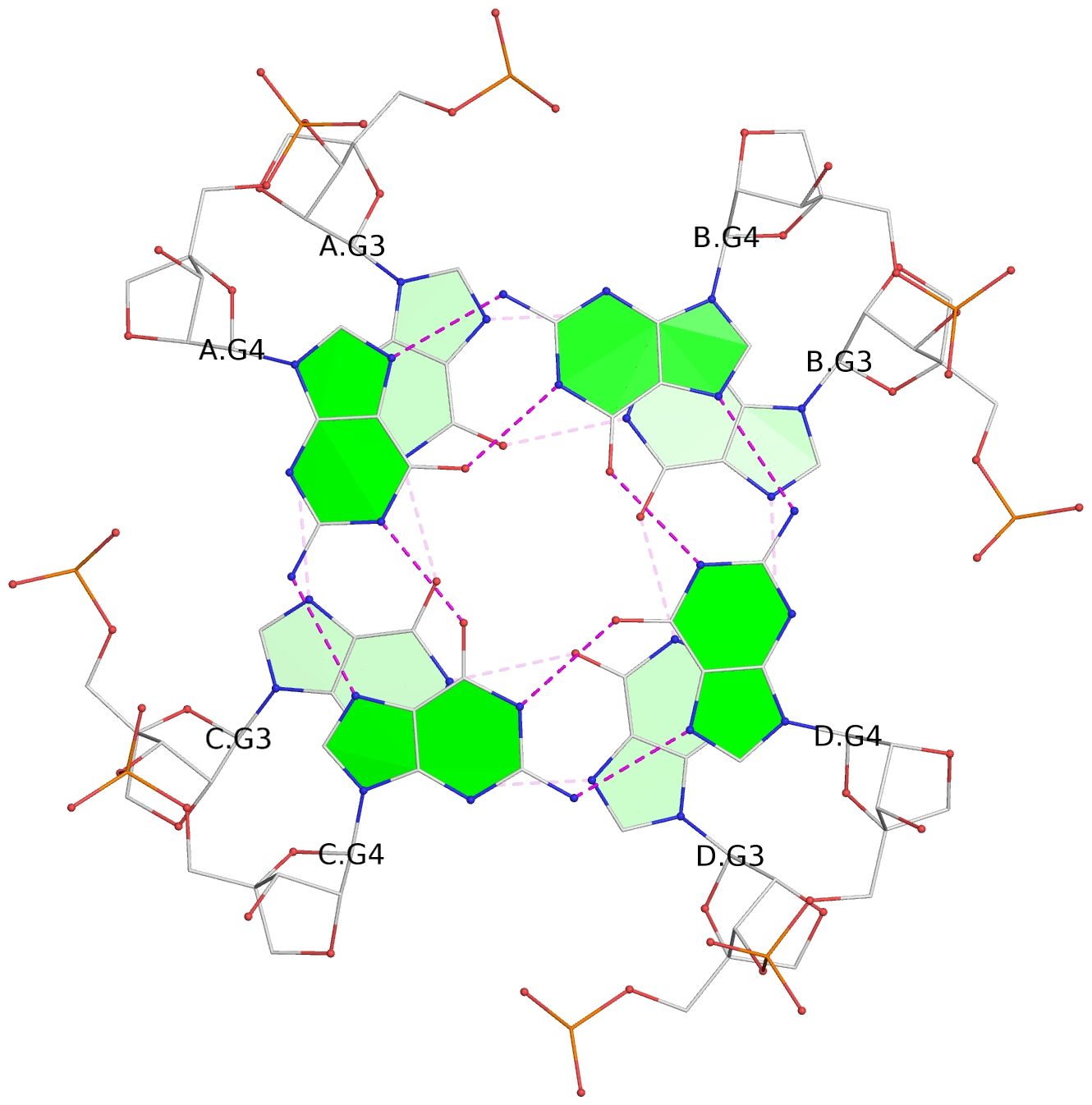
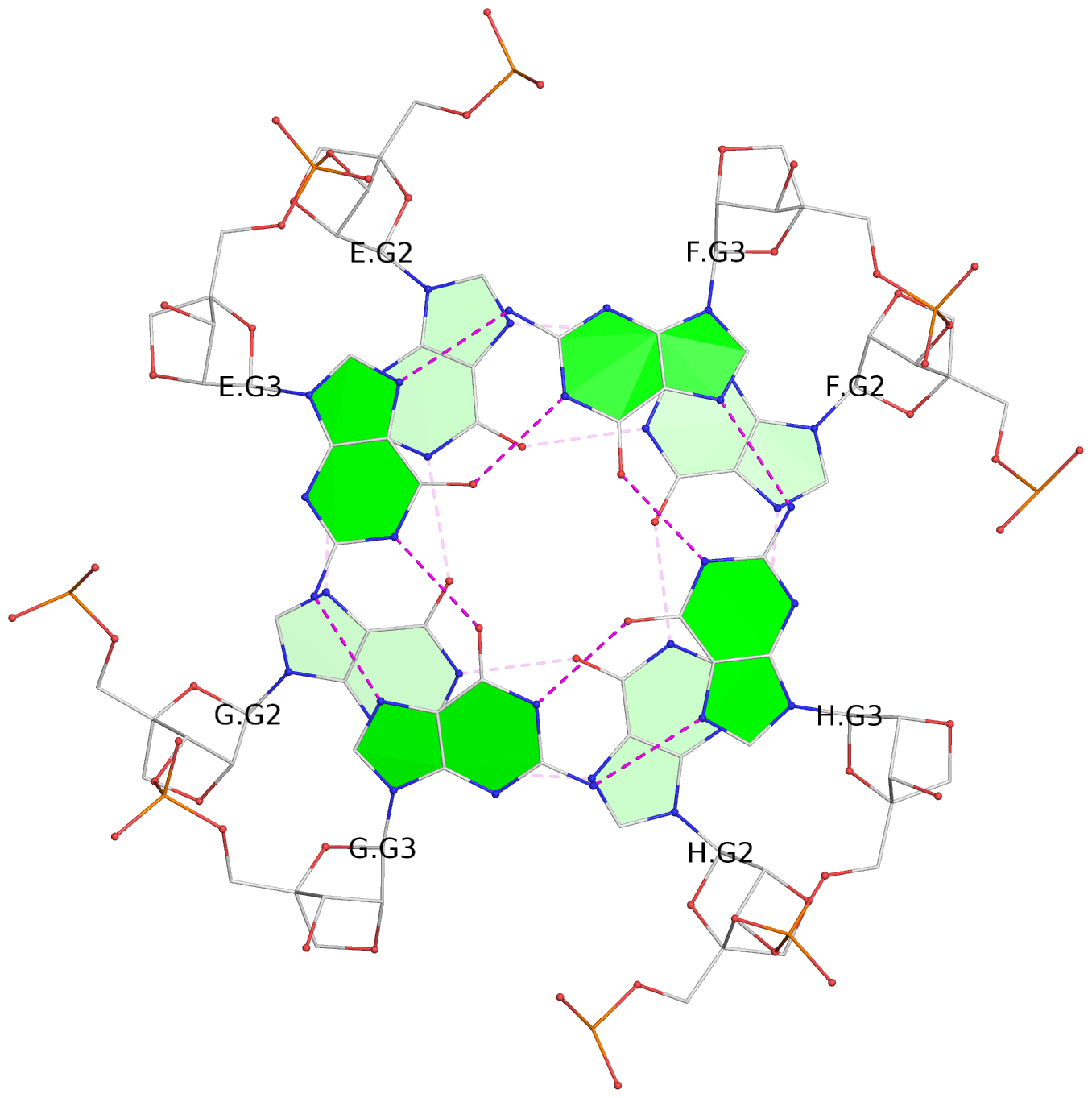
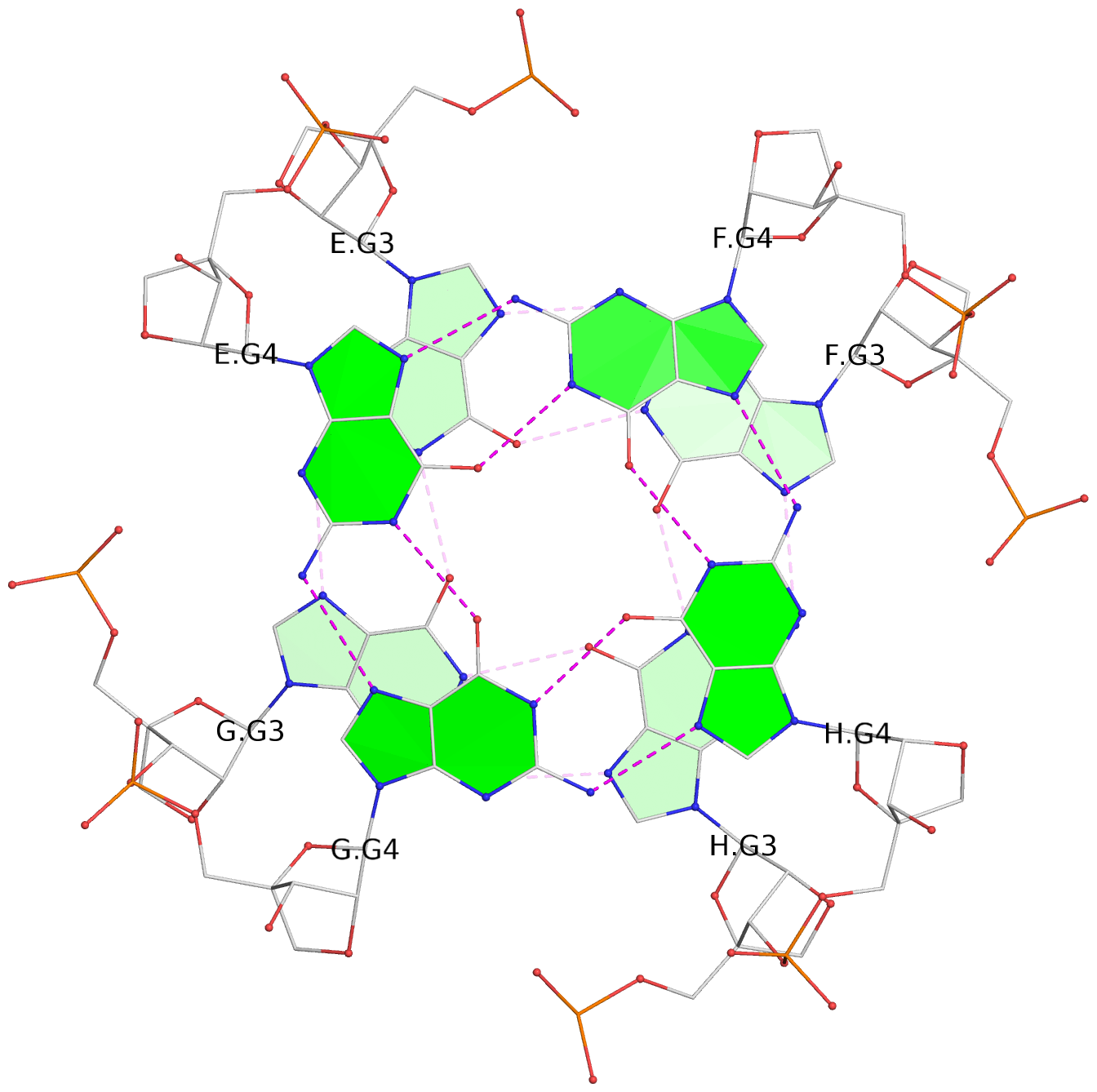
Base-block schematics in six views
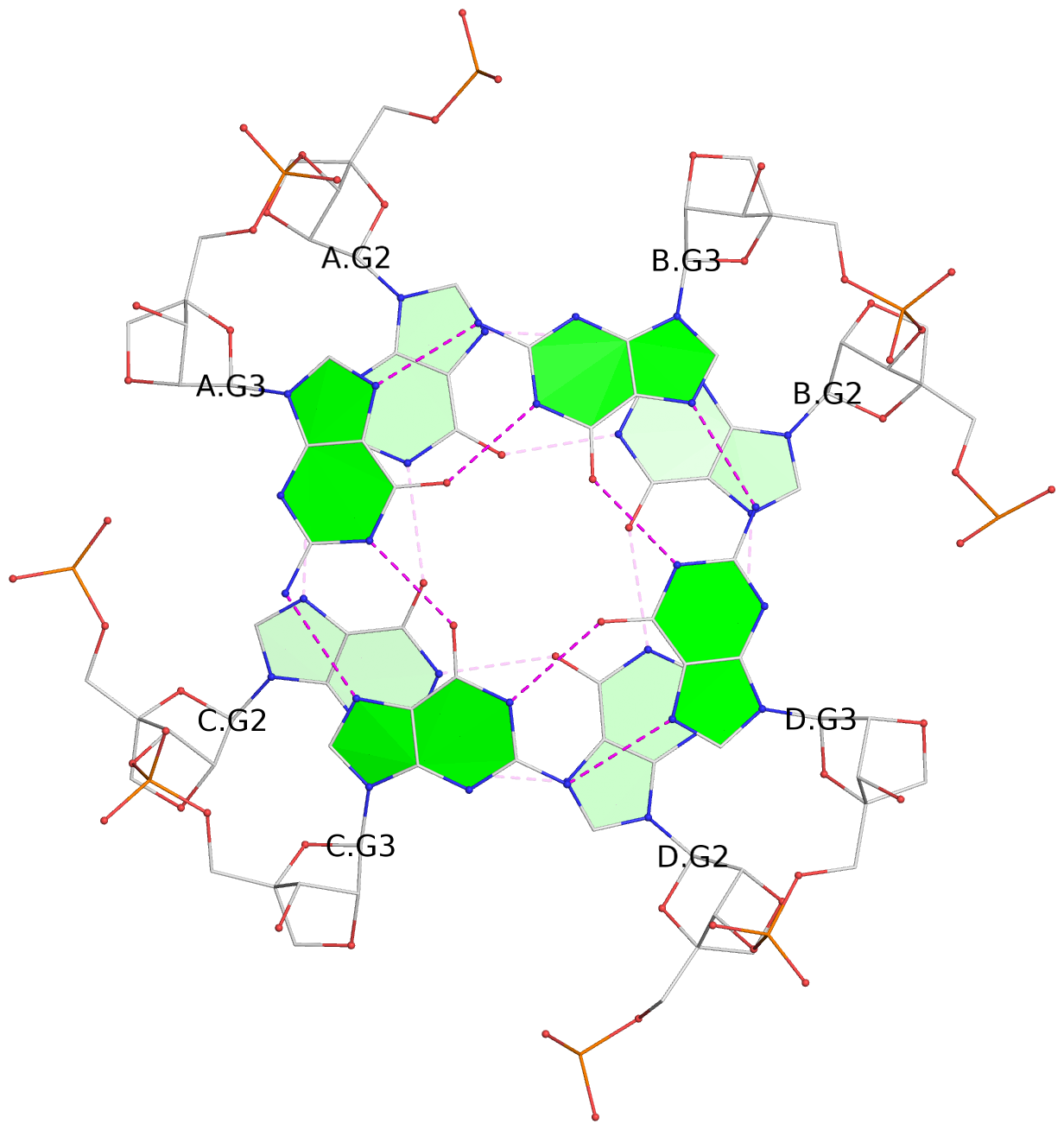
List of 6 G-tetrads
1 glyco-bond=---- sugar=3333 groove=---- planarity=0.420 type=other nts=4 gggg A.LCG2,C.LCG2,D.LCG2,B.LCG2 2 glyco-bond=---- sugar=3333 groove=---- planarity=0.454 type=other nts=4 gggg A.LCG3,C.LCG3,D.LCG3,B.LCG3 3 glyco-bond=---- sugar=3333 groove=---- planarity=0.319 type=other nts=4 gggg A.LCG4,C.LCG4,D.LCG4,B.LCG4 4 glyco-bond=---- sugar=3333 groove=---- planarity=0.434 type=other nts=4 gggg E.LCG2,G.LCG2,H.LCG2,F.LCG2 5 glyco-bond=---- sugar=3333 groove=---- planarity=0.454 type=other nts=4 gggg E.LCG3,G.LCG3,H.LCG3,F.LCG3 6 glyco-bond=---- sugar=3333 groove=---- planarity=0.330 type=other nts=4 gggg E.LCG4,G.LCG4,H.LCG4,F.LCG4
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
Helix#2, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.