Detailed DSSR results for the G-quadruplex: PDB entry 4ni9
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 4ni9
- Class
- cytokine-DNA
- Method
- X-ray (2.55 Å)
- Summary
- Crystal structure of human interleukin 6 in complex with a modified nucleotide aptamer (somamer sl1025), form 2
- Reference
- Gelinas AD, Davies DR, Edwards TE, Rohloff JC, Carter JD, Zhang C, Gupta S, Ishikawa Y, Hirota M, Nakaishi Y, Jarvis TC, Janjic N (2014): "Crystal structure of interleukin-6 in complex with a modified nucleic Acid ligand." J.Biol.Chem., 289, 8720-8734. doi: 10.1074/jbc.M113.532697.
- Abstract
- IL-6 is a secreted cytokine that functions through binding two cell surface receptors, IL-6Rα and gp130. Because of its involvement in the progression of several chronic inflammatory diseases, IL-6 is a target of pharmacologic interest. We have recently identified a novel class of ligands called SOMAmers (S low Off-rate Modified Aptamers) that bind IL-6 and inhibit its biologic activity. SOMAmers exploit the chemical diversity of protein-like side chains assembled on flexible nucleic acid scaffolds, resulting in an expanded repertoire of intra- and intermolecular interactions not achievable with conventional aptamers. Here, we report the co-crystal structure of a high affinity SOMAmer (Kd = 0.20 nm) modified at the 5-position of deoxyuridine in a complex with IL-6. The SOMAmer, comprised of a G-quartet domain and a stem-loop domain, engages IL-6 in a clamp-like manner over an extended surface exhibiting close shape complementarity with the protein. The interface is characterized by substantial hydrophobic interactions overlapping the binding surfaces of the IL-6Rα and gp130 receptors. The G-quartet domain retains considerable binding activity as a disconnected autonomous fragment (Kd = 270 nm). A single substitution from our diversely modified nucleotide library leads to a 37-fold enhancement in binding affinity of the G-quartet fragment (Kd = 7.4 nm). The ability to probe ligand surfaces in this manner is a powerful tool in the development of new therapeutic reagents with improved pharmacologic properties. The SOMAmer·IL-6 structure also expands our understanding of the diverse structural motifs achievable with modified nucleic acid libraries and elucidates the nature with which these unique ligands interact with their protein targets.
- G4 notes
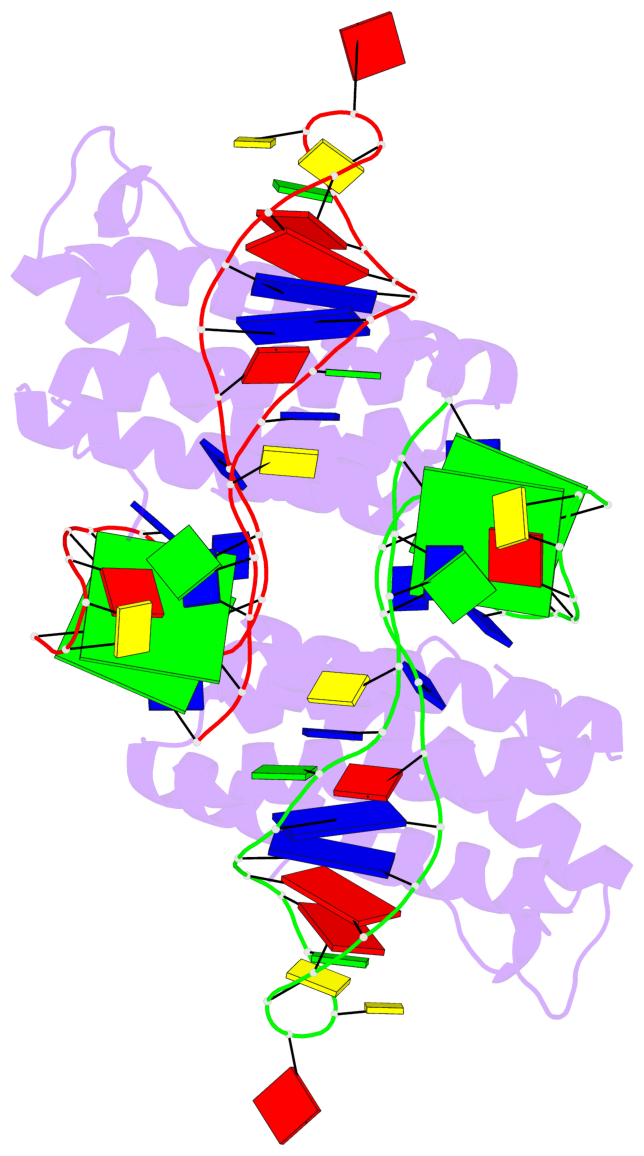
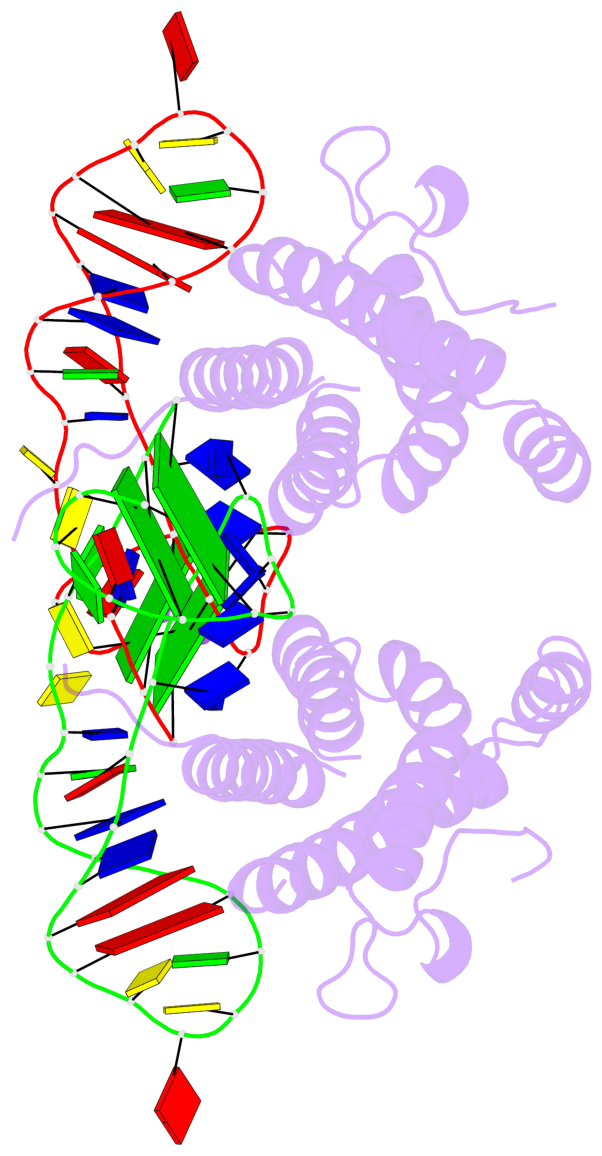
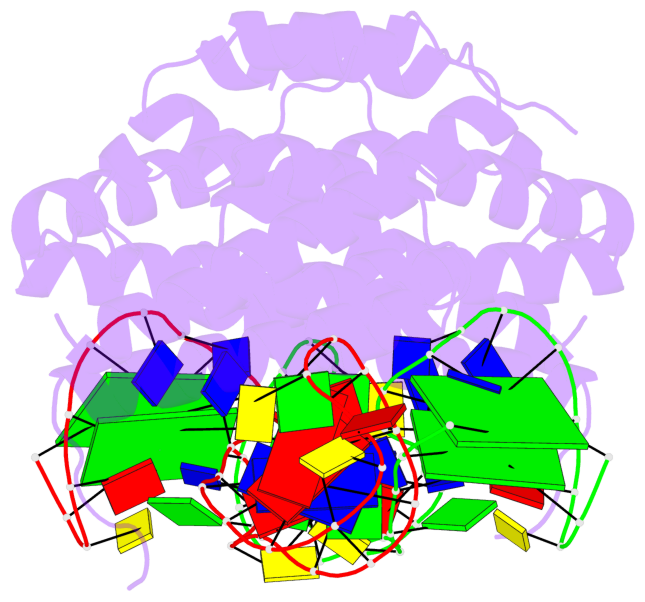
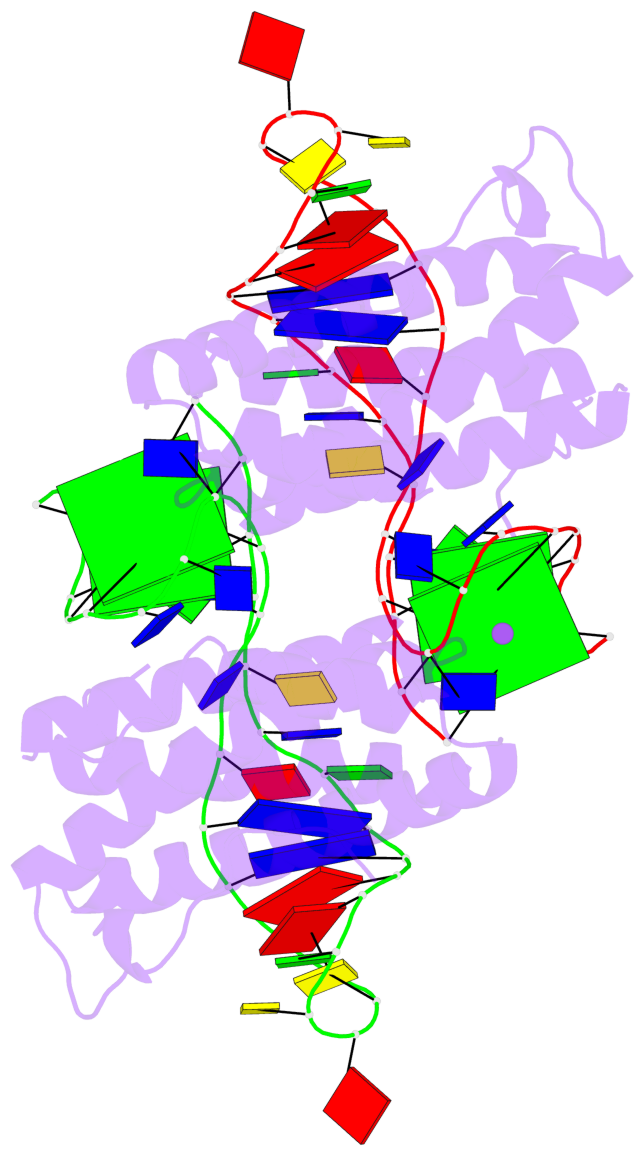
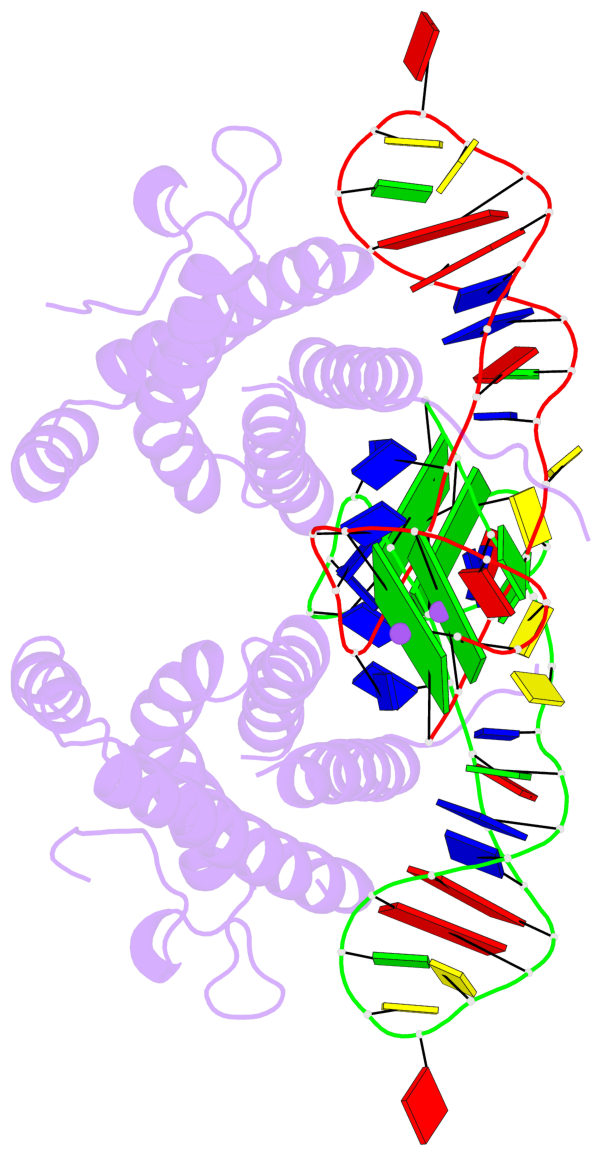
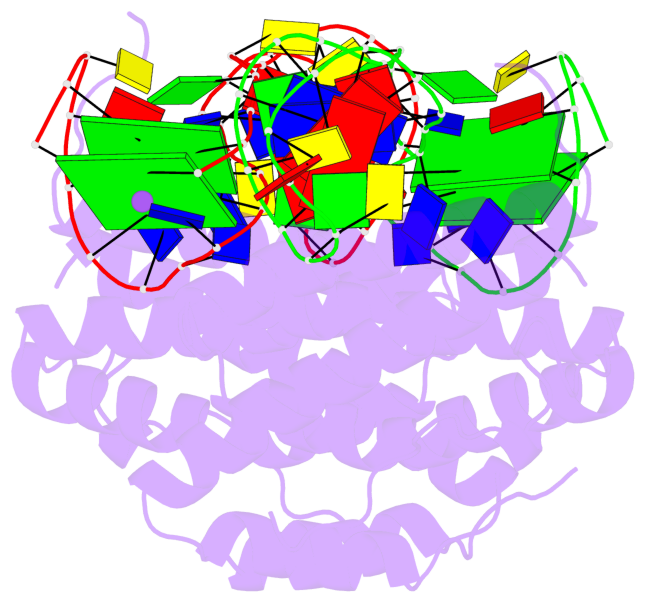
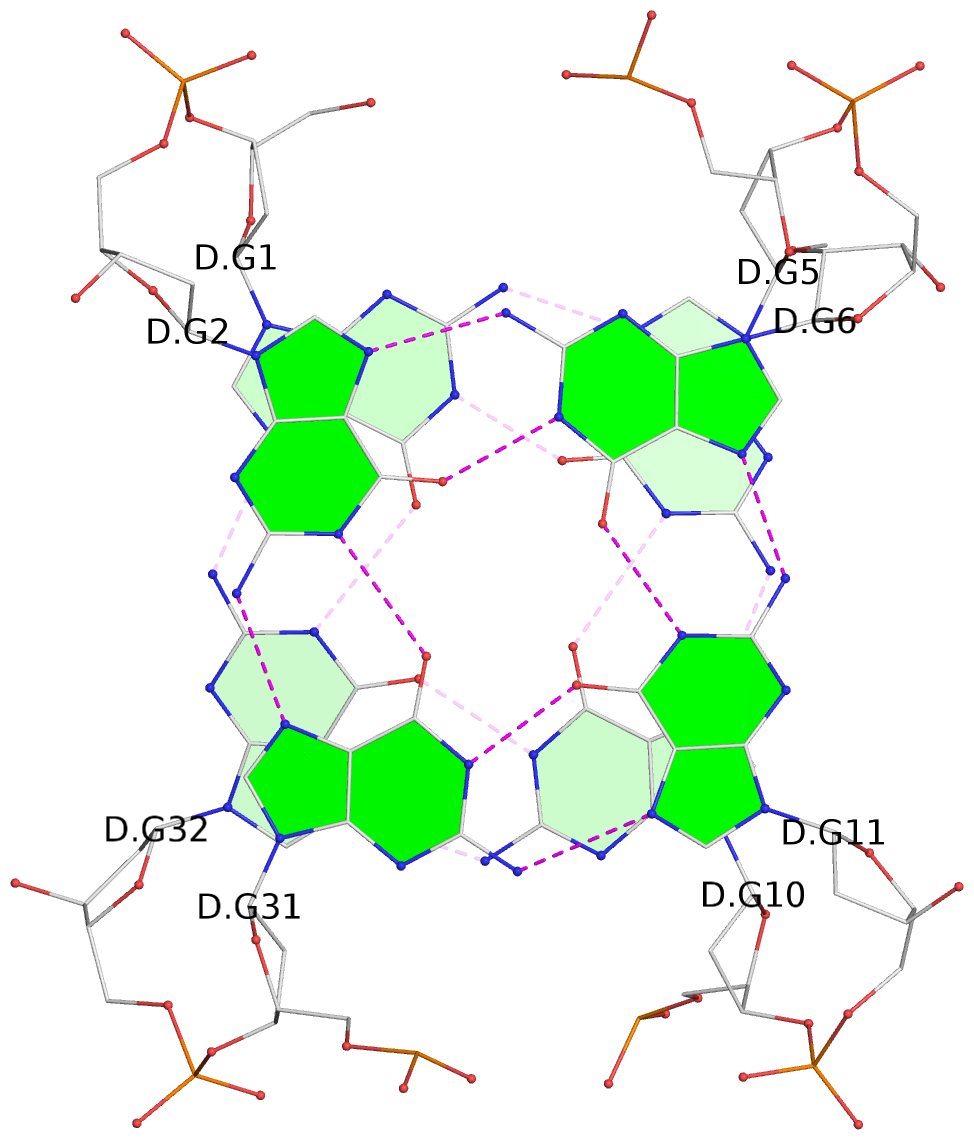
- 4 G-tetrads, 2 G4 helices, 2 G4 stems, 2(+Ln+Lw+Ln), chair(2+2), UDUD
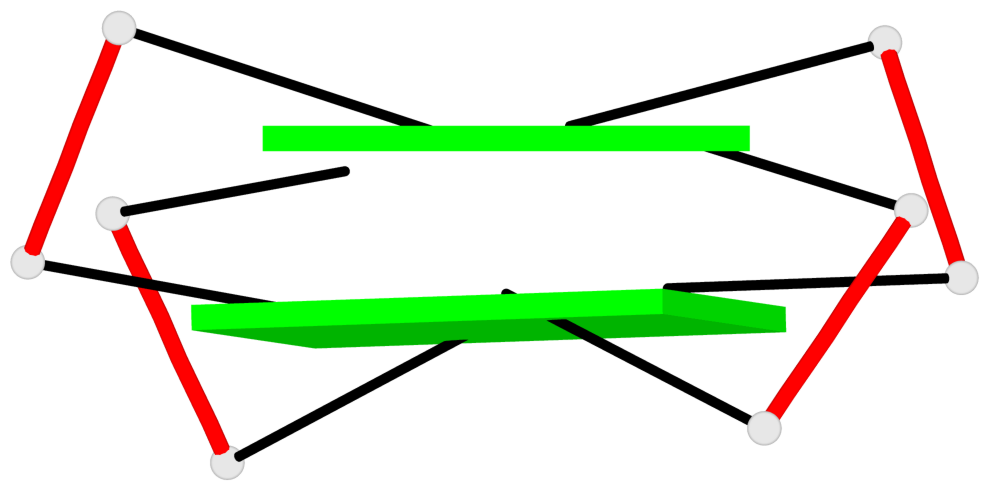
Base-block schematics in six views
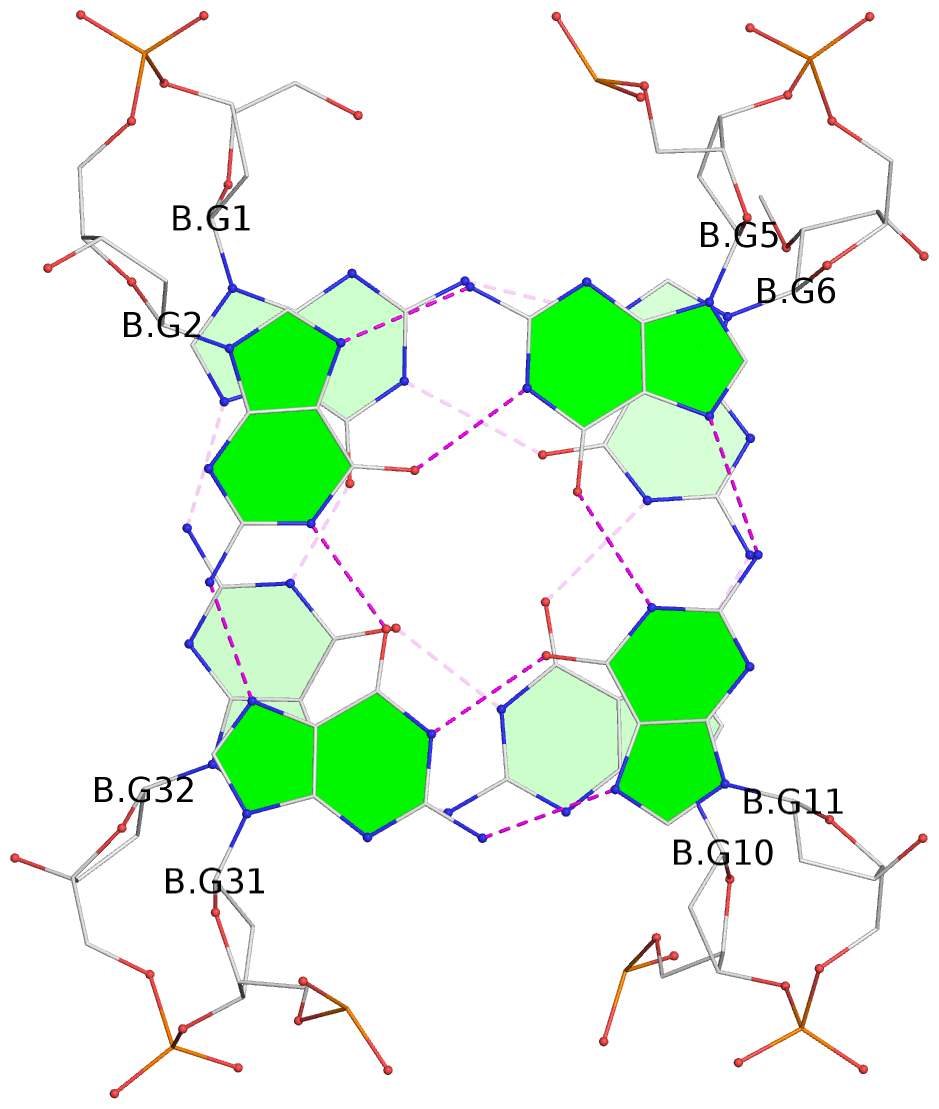
List of 4 G-tetrads
1 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.693 type=other nts=4 GGGg B.DG1,B.DG32,B.DG10,B.OMG6 2 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.718 type=bowl nts=4 GGGG B.DG2,B.DG31,B.DG11,B.DG5 3 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.672 type=other nts=4 GGGg D.DG1,D.DG32,D.DG10,D.OMG6 4 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.625 type=bowl-2 nts=4 GGGG D.DG2,D.DG31,D.DG11,D.DG5
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular, with 1 stem
Helix#2, 2 G-tetrad layers, INTRA-molecular, with 1 stem
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.