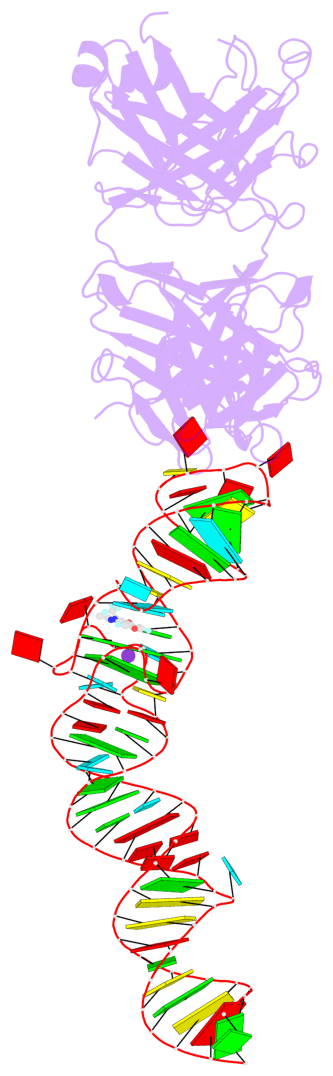
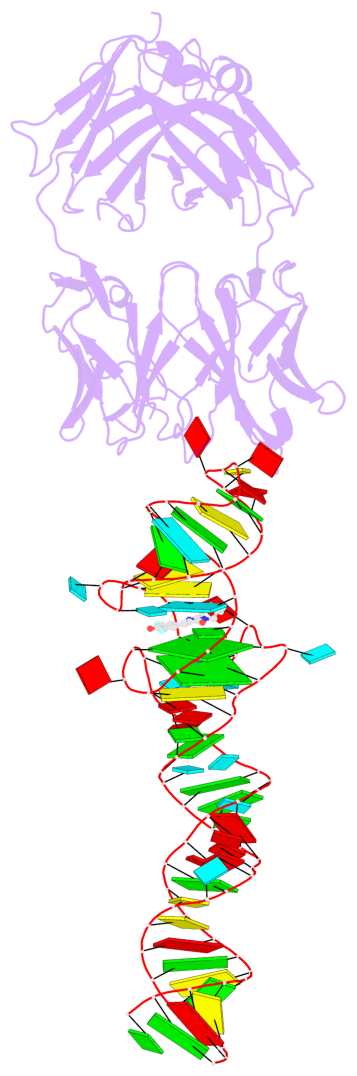
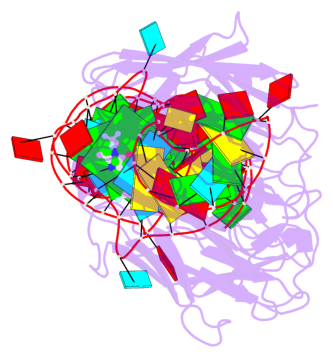
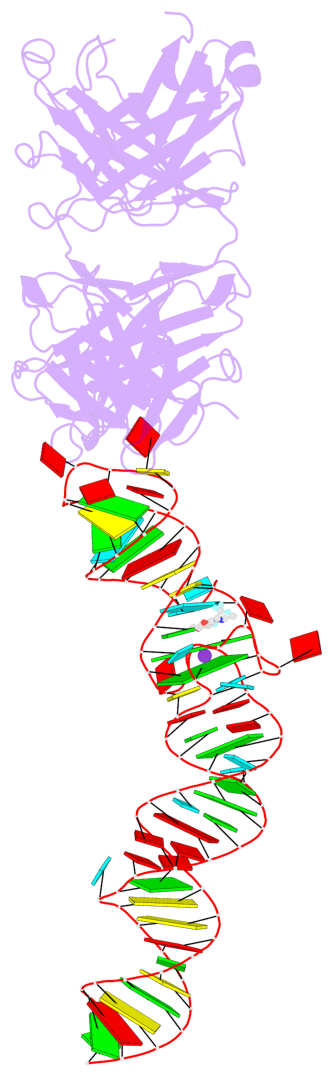
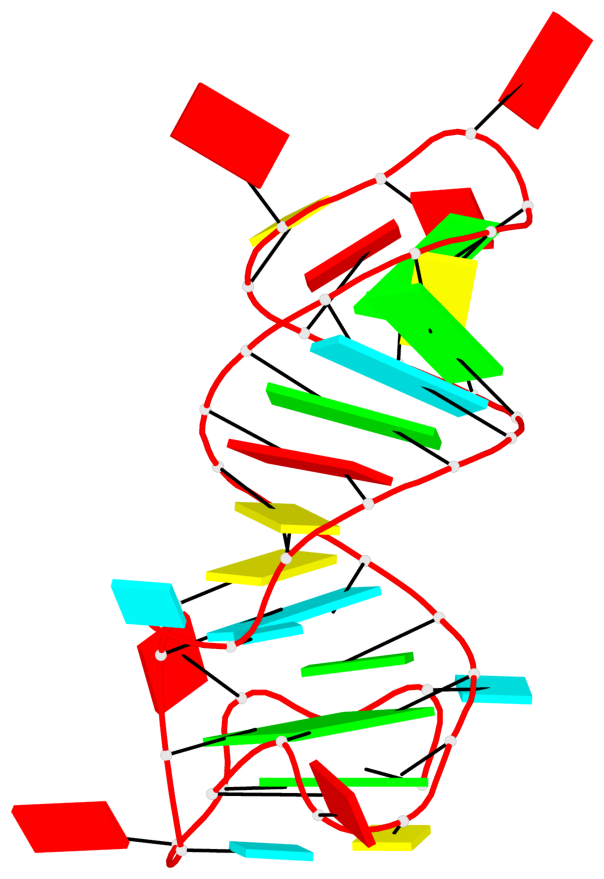
Detailed DSSR results for the G-quadruplex: PDB entry 4q9r
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 4q9r
- Class
- RNA-immune system
- Method
- X-ray (3.12 Å)
- Summary
- Crystal structure of an RNA aptamer bound to trifluoroethyl-ligand analog in complex with fab
- Reference
- Huang H, Suslov NB, Li NS, Shelke SA, Evans ME, Koldobskaya Y, Rice PA, Piccirilli JA (2014): "A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore." Nat.Chem.Biol., 10, 686-691. doi: 10.1038/nchembio.1561.
- Abstract
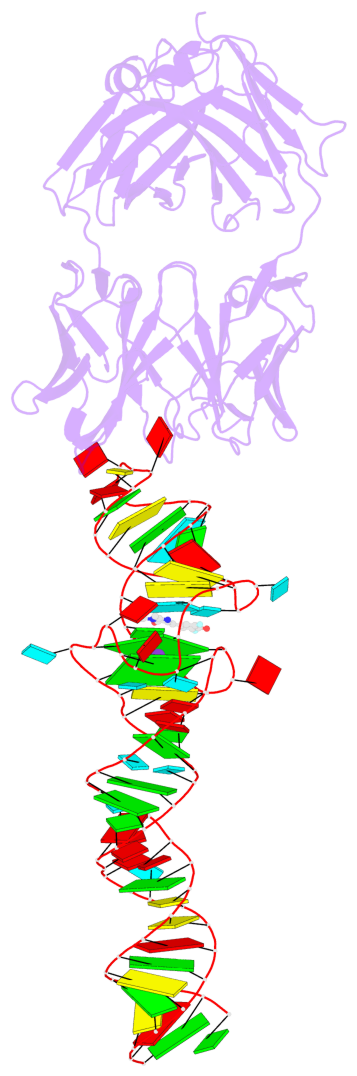
- Spinach is an in vitro-selected RNA aptamer that binds a GFP-like ligand and activates its green fluorescence. Spinach is thus an RNA analog of GFP and has potentially widespread applications for in vivo labeling and imaging. We used antibody-assisted crystallography to determine the structures of Spinach both with and without bound fluorophore at 2.2-Å and 2.4-Å resolution, respectively. Spinach RNA has an elongated structure containing two helical domains separated by an internal bulge that folds into a G-quadruplex motif of unusual topology. The G-quadruplex motif and adjacent nucleotides comprise a partially preformed binding site for the fluorophore. The fluorophore binds in a planar conformation and makes extensive aromatic stacking and hydrogen bond interactions with the RNA. Our findings provide a foundation for structure-based engineering of new fluorophore-binding RNA aptamers.
- G4 notes
- 2 G-tetrads, 1 G4 helix, 1 G4 stem, 2(-PD+P), (2+2), UUDD
Base-block schematics in six views
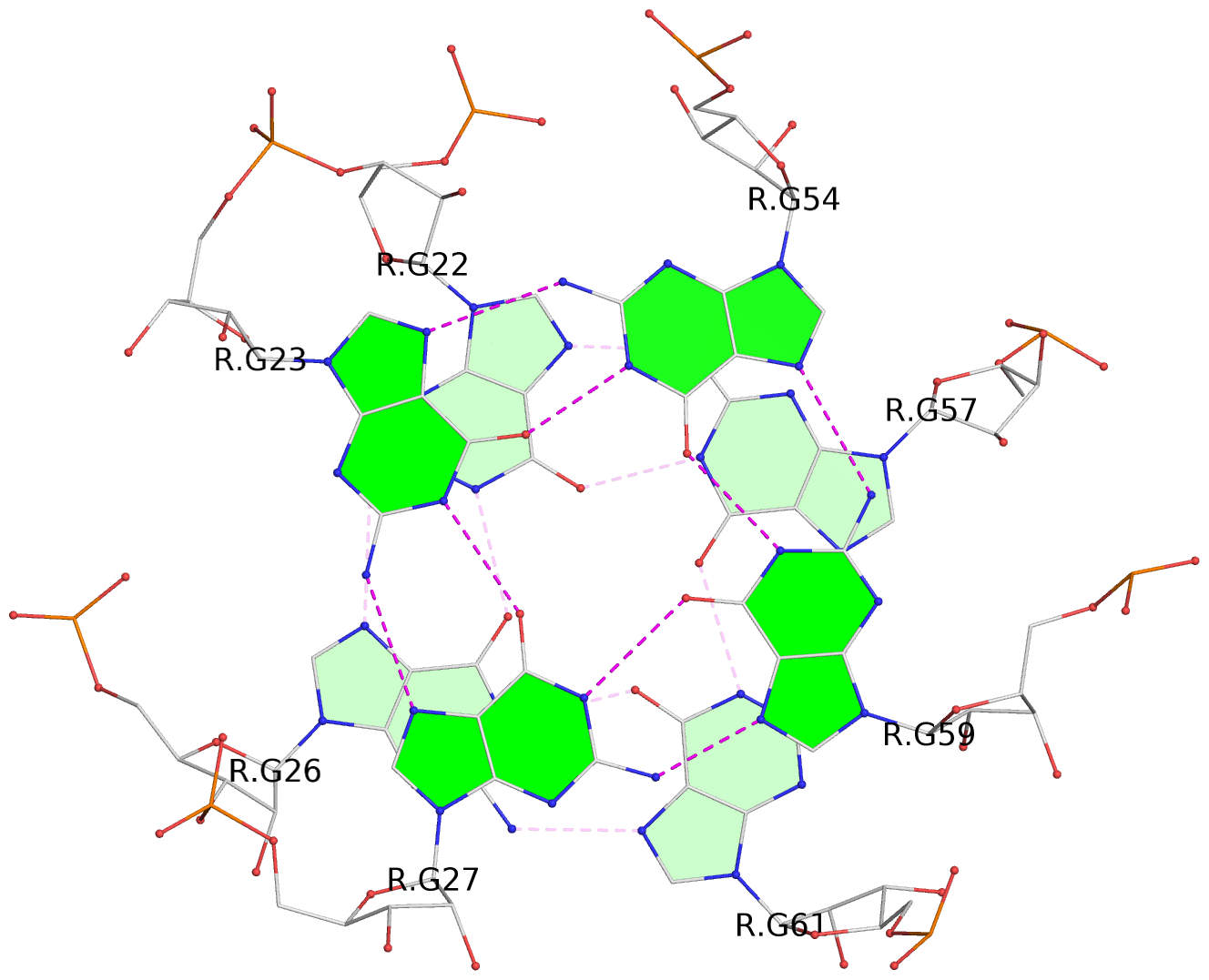
List of 2 G-tetrads
1 glyco-bond=--s- sugar=-33- groove=-wn- planarity=0.268 type=other nts=4 GGGG R.G22,R.G26,R.G61,R.G57 2 glyco-bond=--ss sugar=-3-3 groove=-w-n planarity=0.128 type=planar nts=4 GGGG R.G23,R.G27,R.G59,R.G54
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.