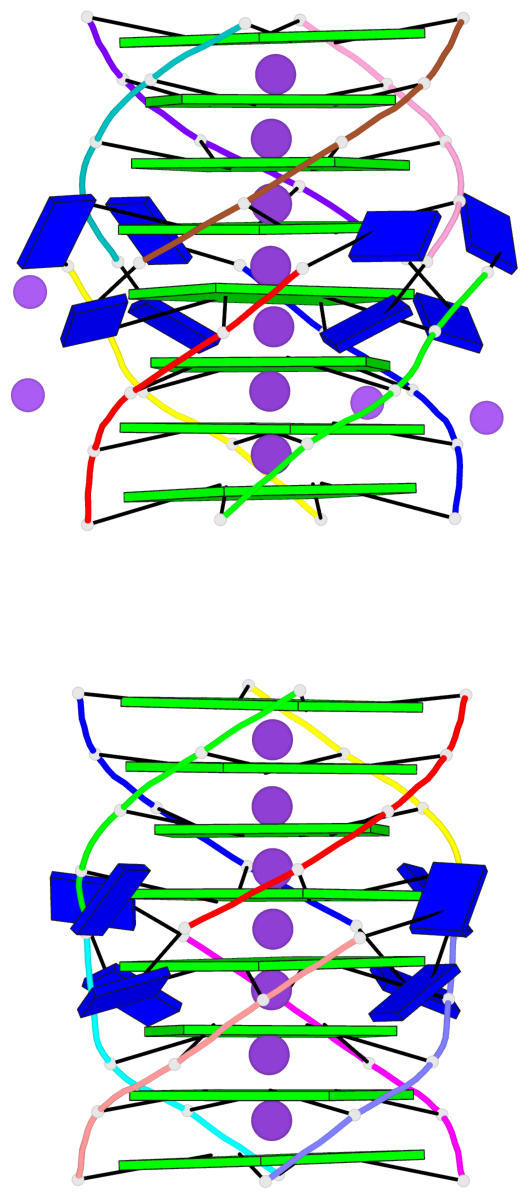
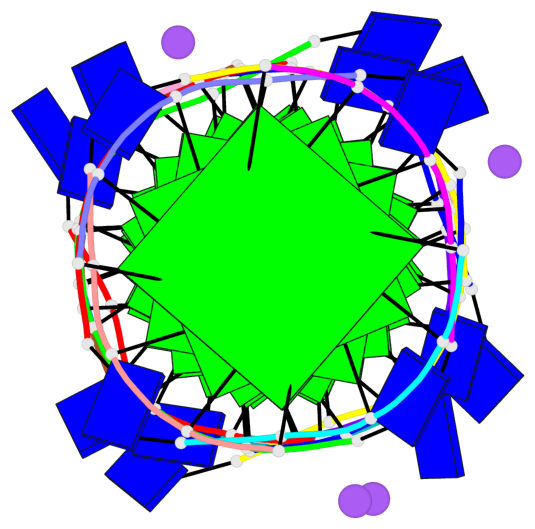
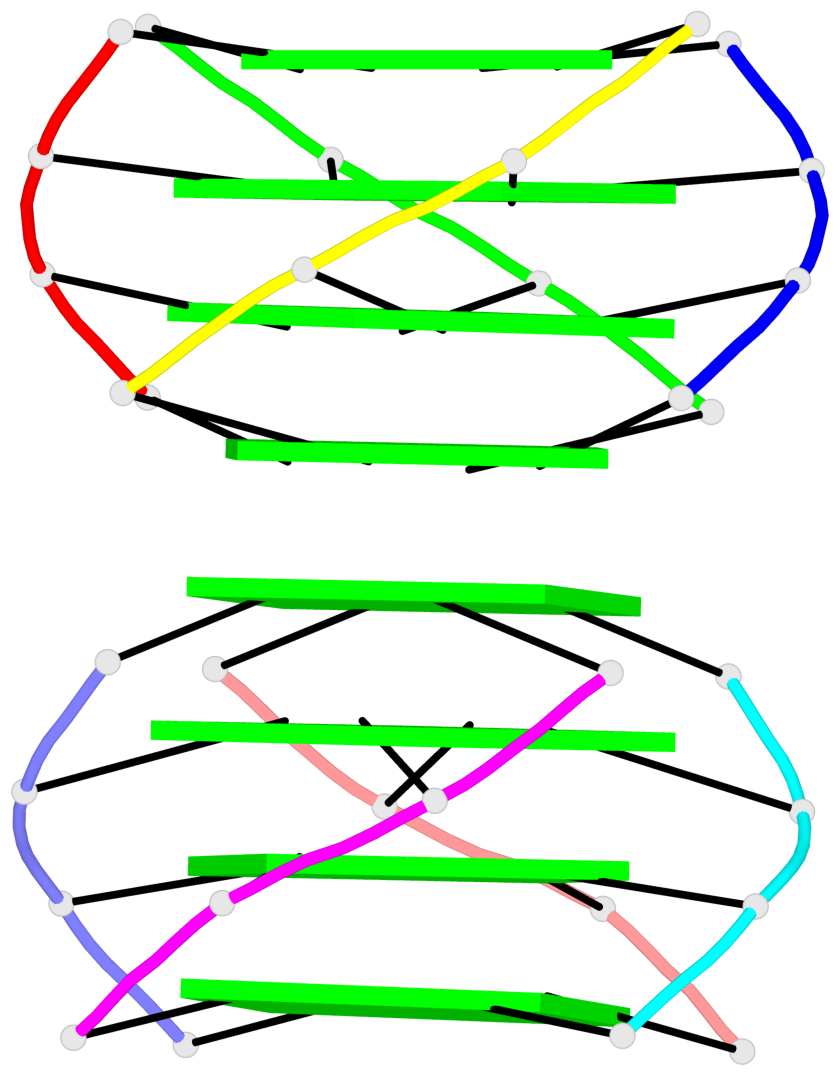
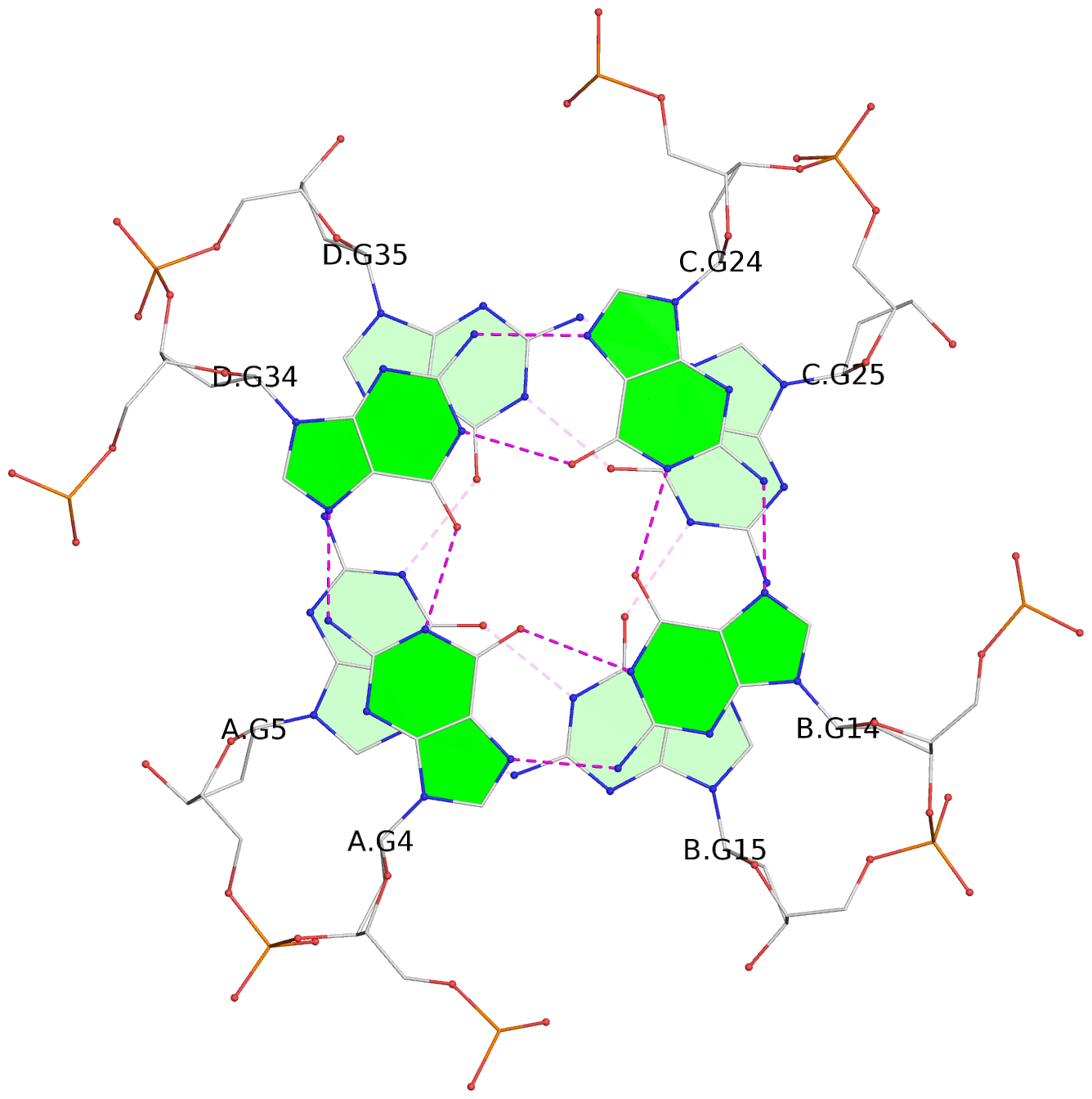
Detailed DSSR results for the G-quadruplex: PDB entry 4r44
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 4r44
- Class
- DNA
- Method
- X-ray (2.695 Å)
- Summary
- Racemic crystal structure of a tetramolecular DNA g-quadruplex
- Reference
- Mandal PK, Collie GW, Kauffmann B, Huc I (2014): "Racemic DNA crystallography." Angew.Chem.Int.Ed.Engl., 53, 14424-14427. doi: 10.1002/anie.201409014.
- Abstract
- Racemates increase the chances of crystallization by allowing molecular contacts to be formed in a greater number of ways. With the advent of protein synthesis, the production of protein racemates and racemic-protein crystallography are now possible. Curiously, racemic DNA crystallography had not been investigated despite the commercial availability of L- and D-deoxyribo-oligonucleotides. Here, we report a study into racemic DNA crystallography showing the strong propensity of racemic DNA mixtures to form racemic crystals. We describe racemic crystal structures of various DNA sequences and folded conformations, including duplexes, quadruplexes, and a four-way junction, showing that the advantages of racemic crystallography should extend to DNA.
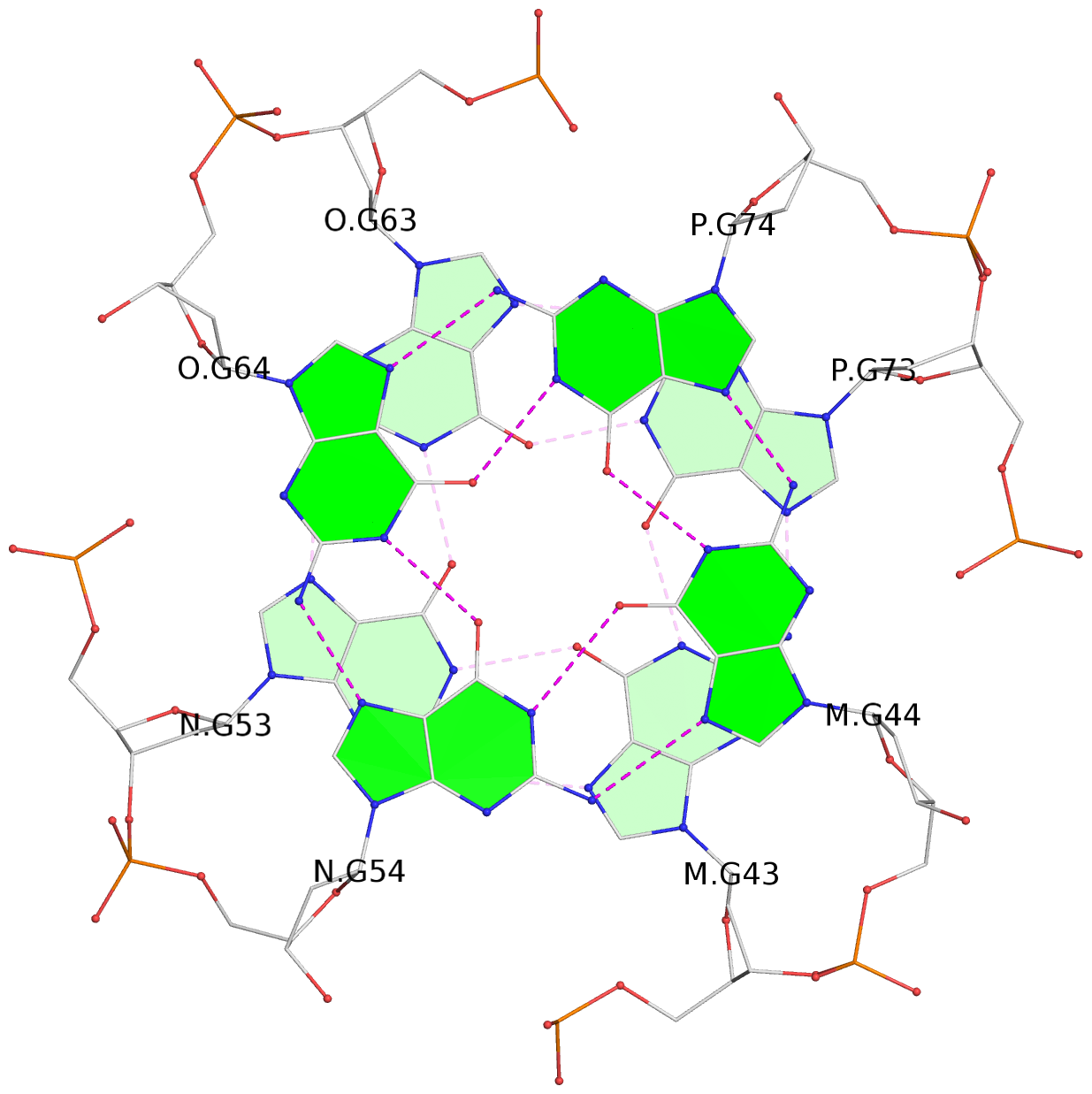
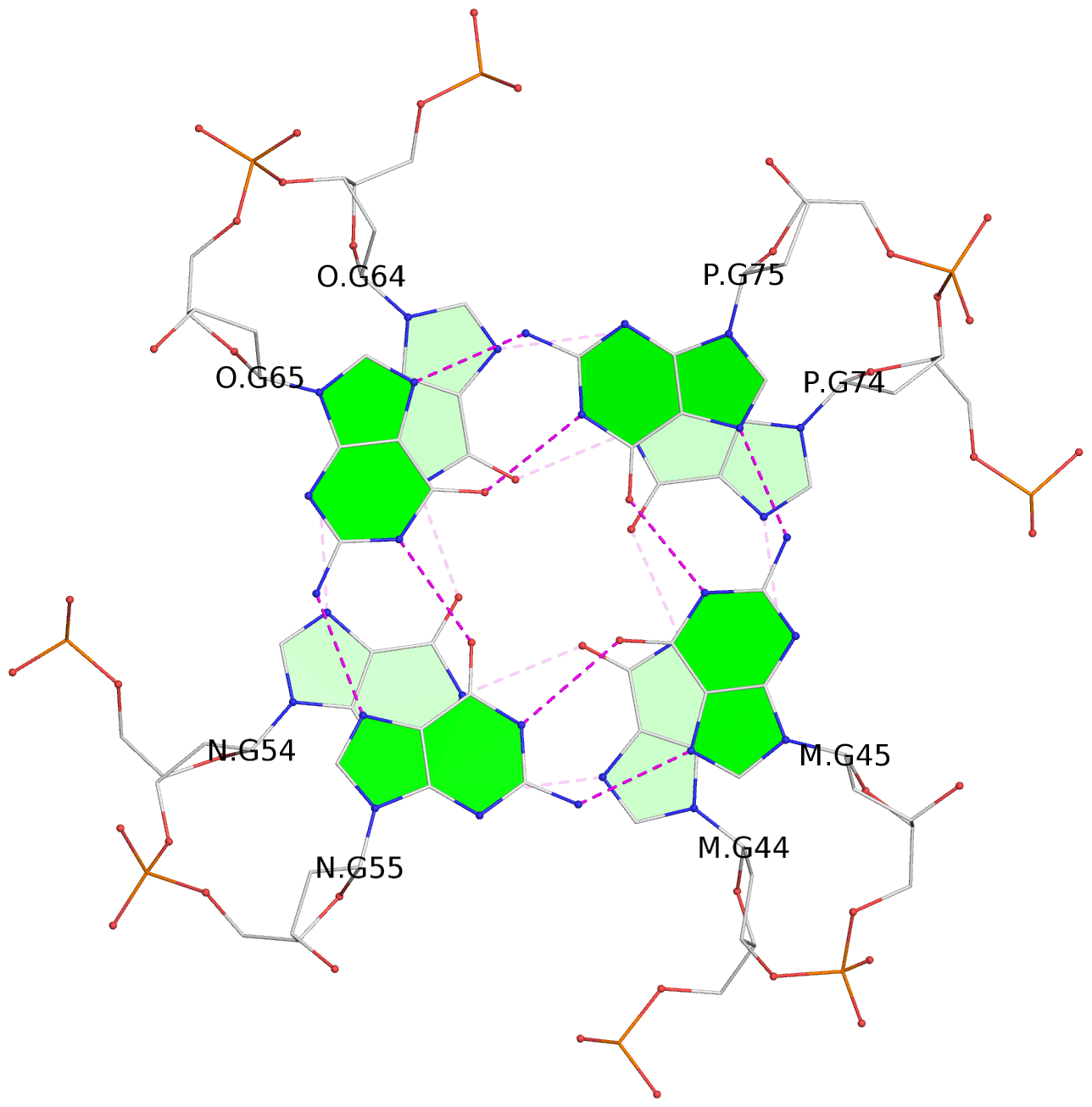
- G4 notes
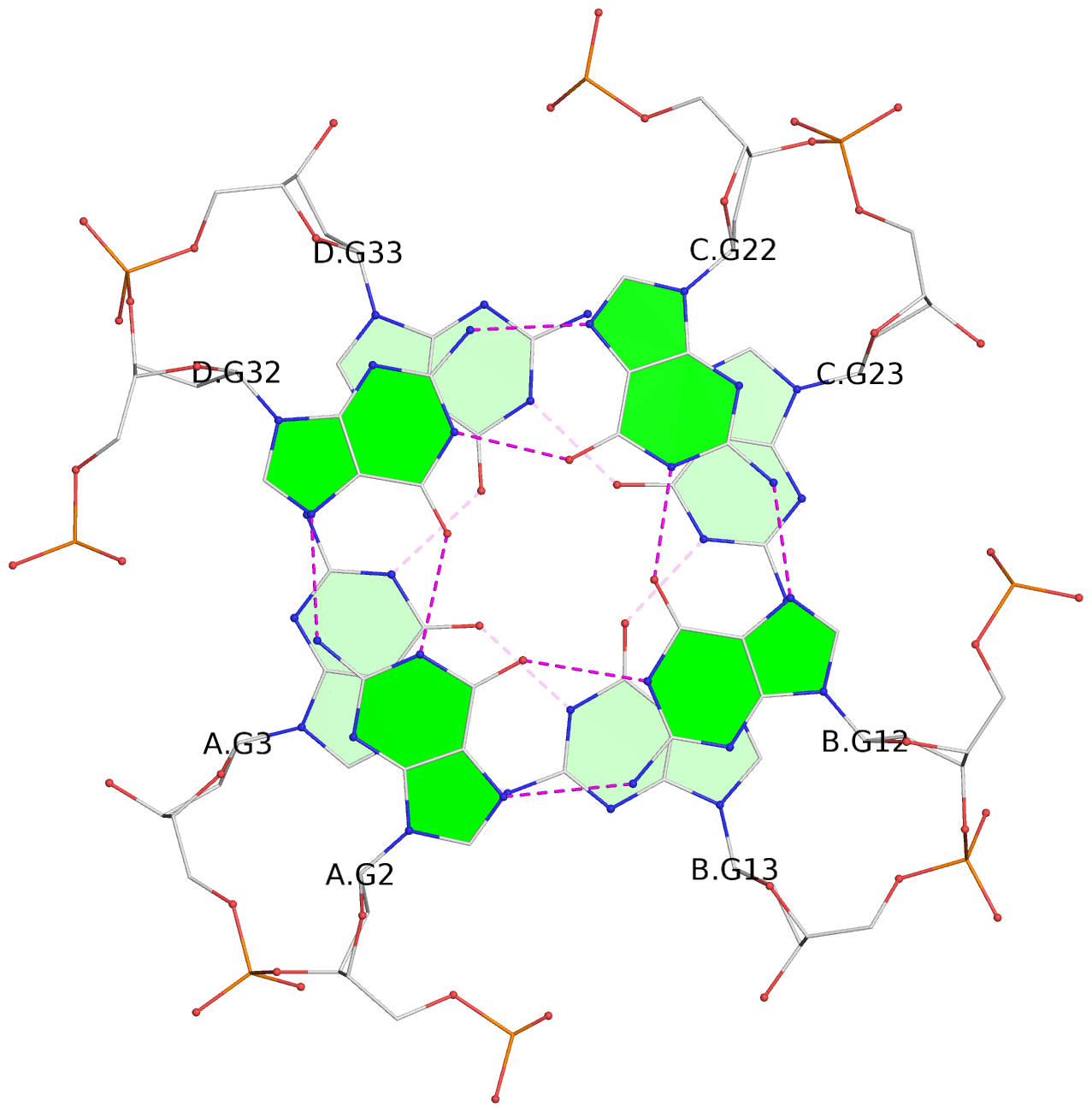
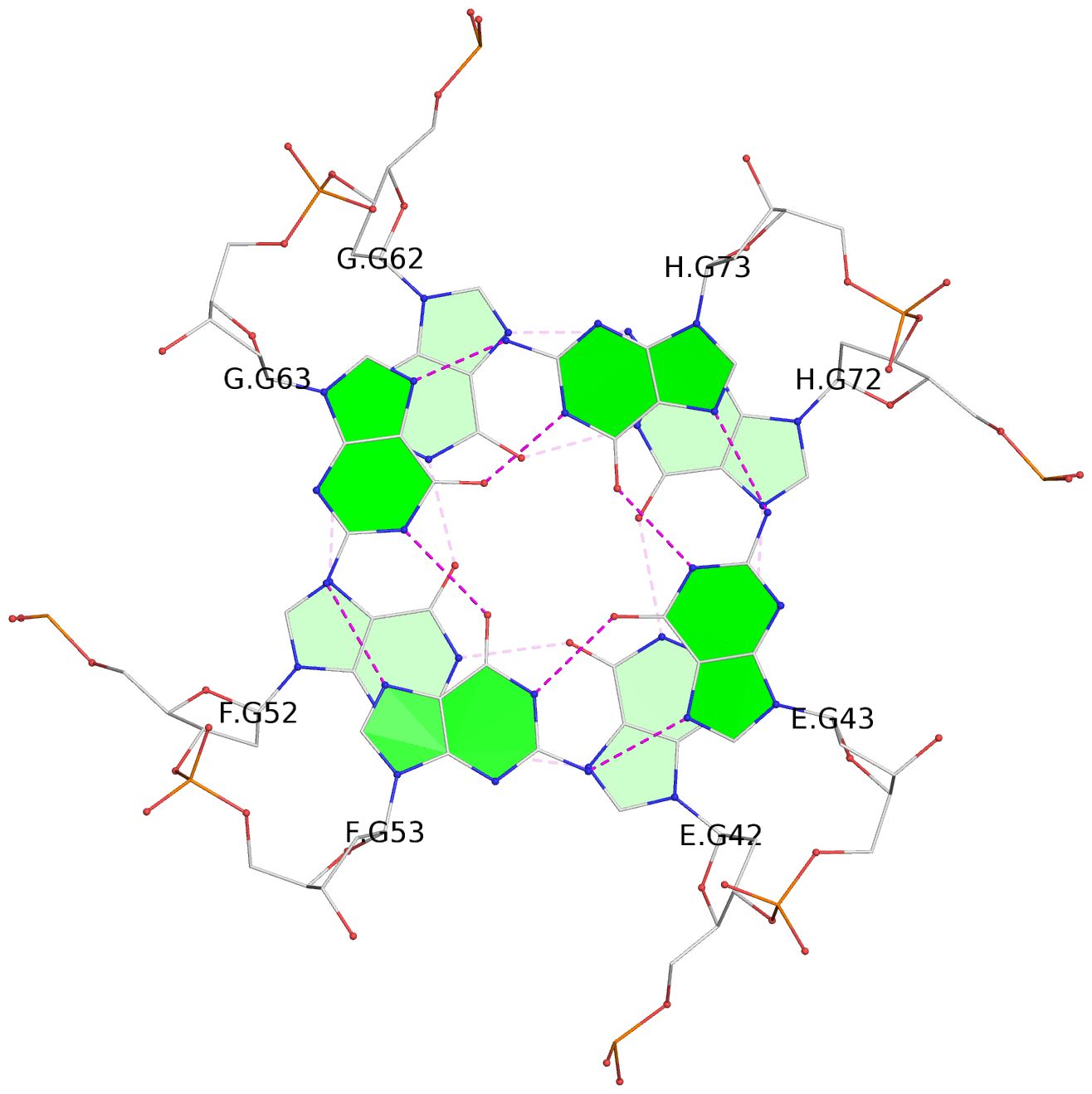
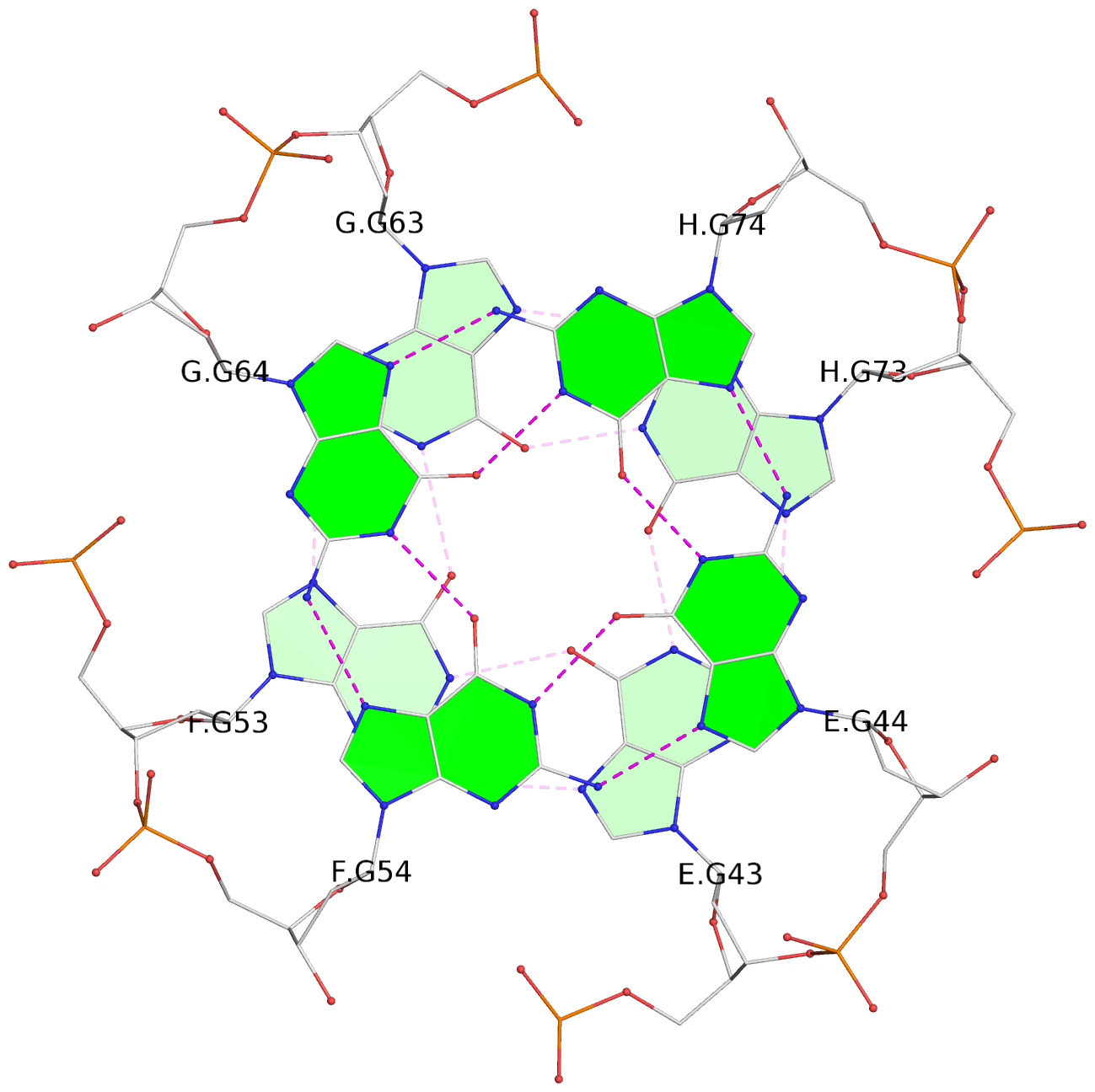
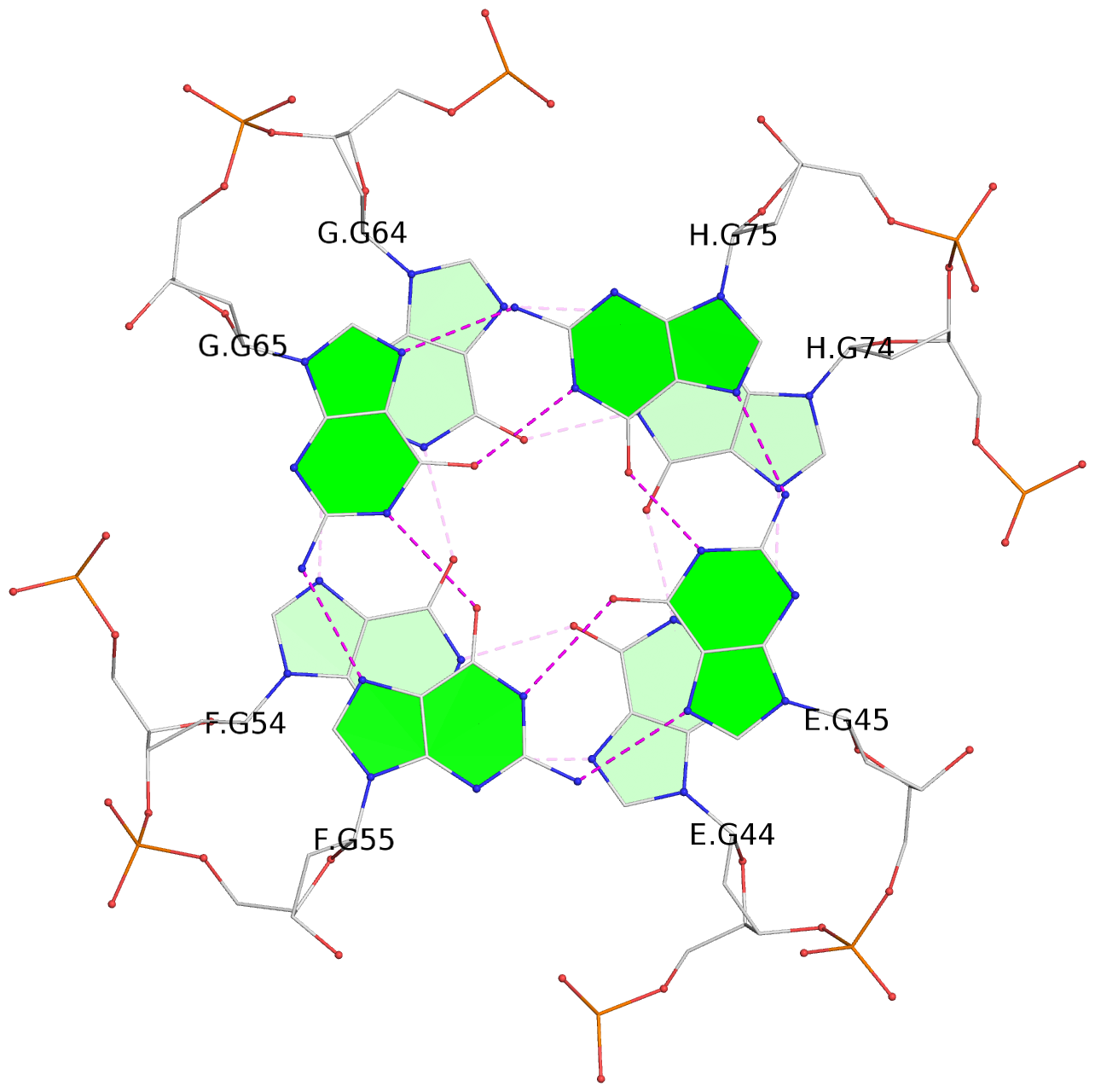
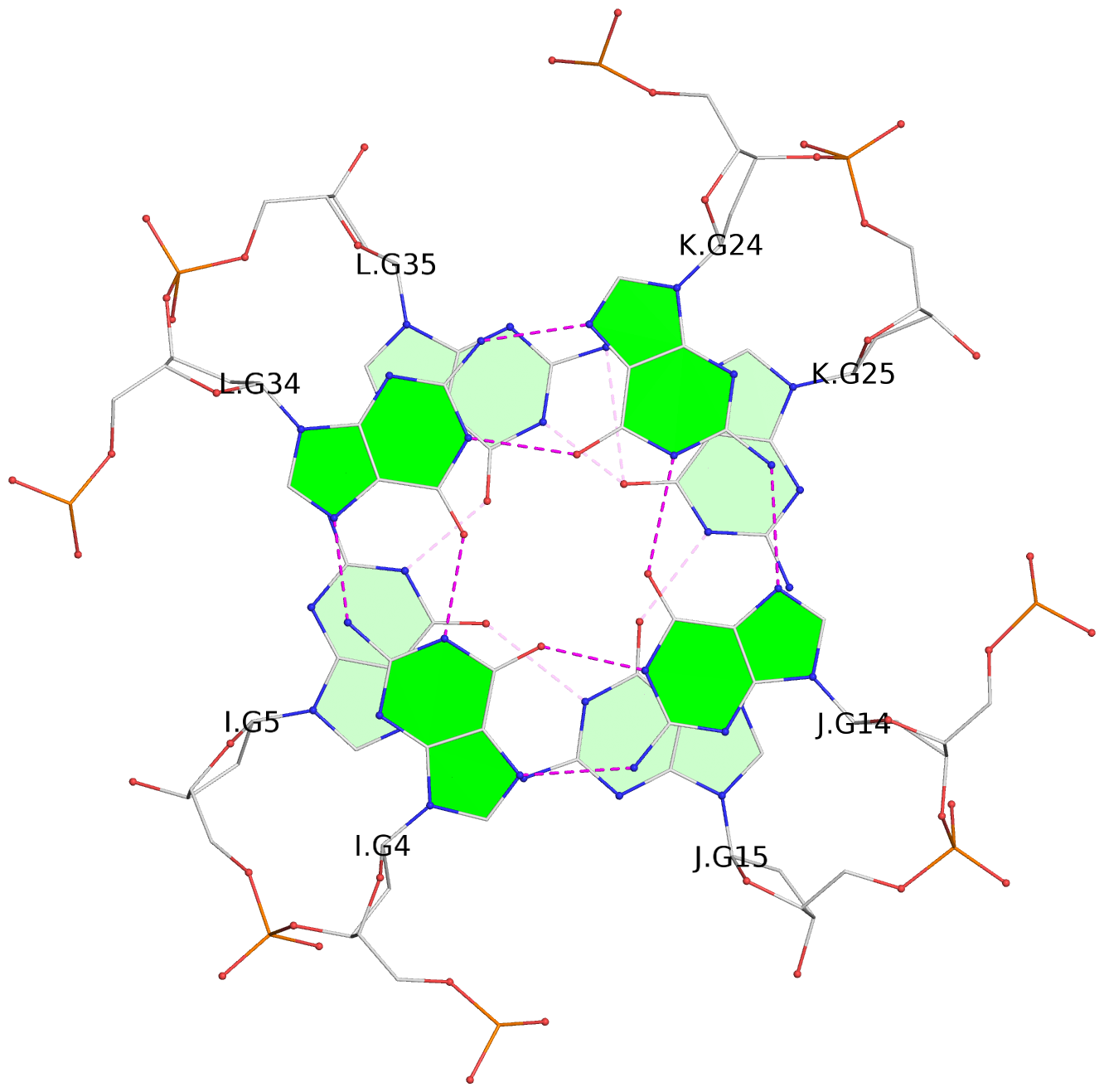
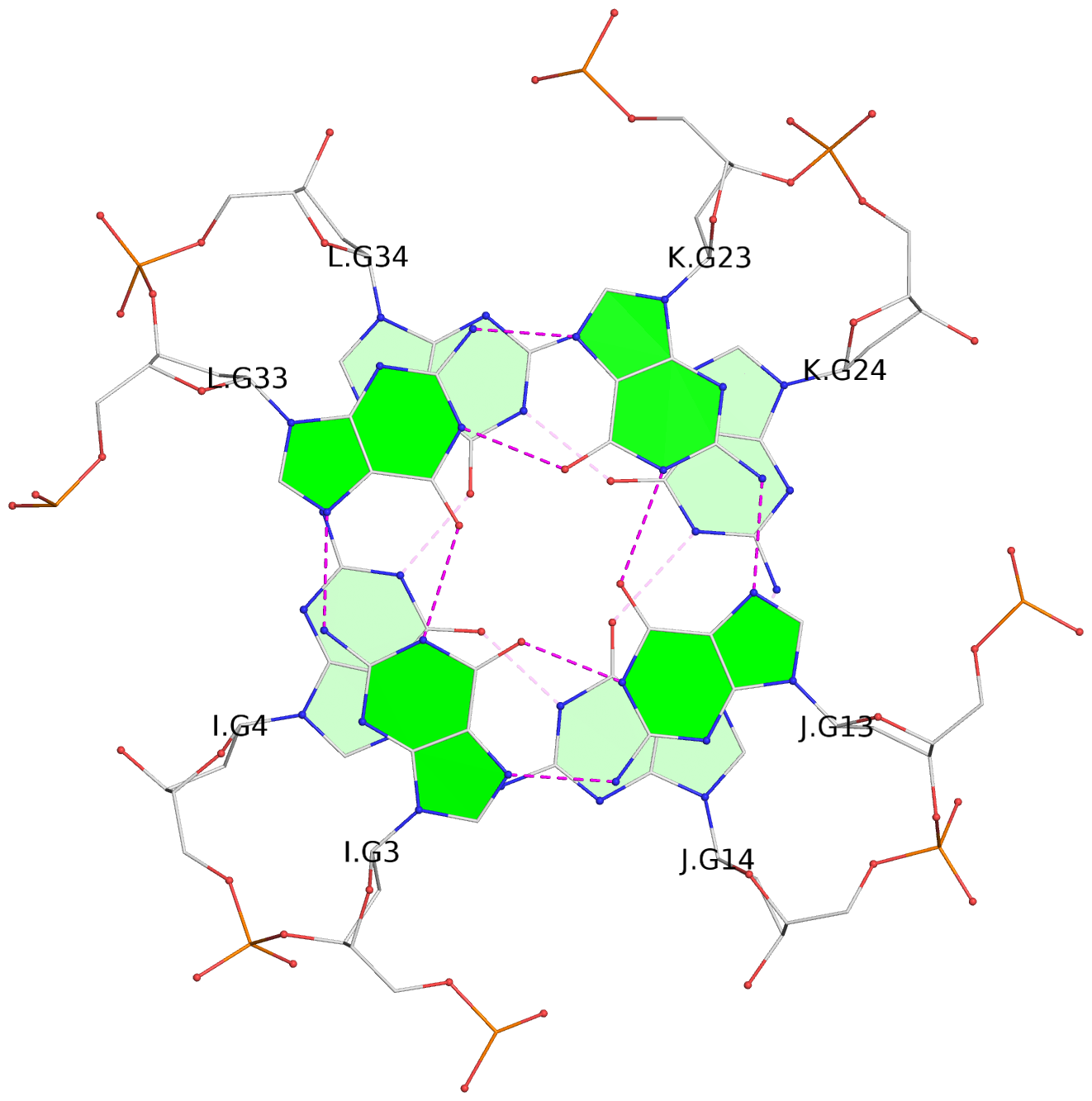
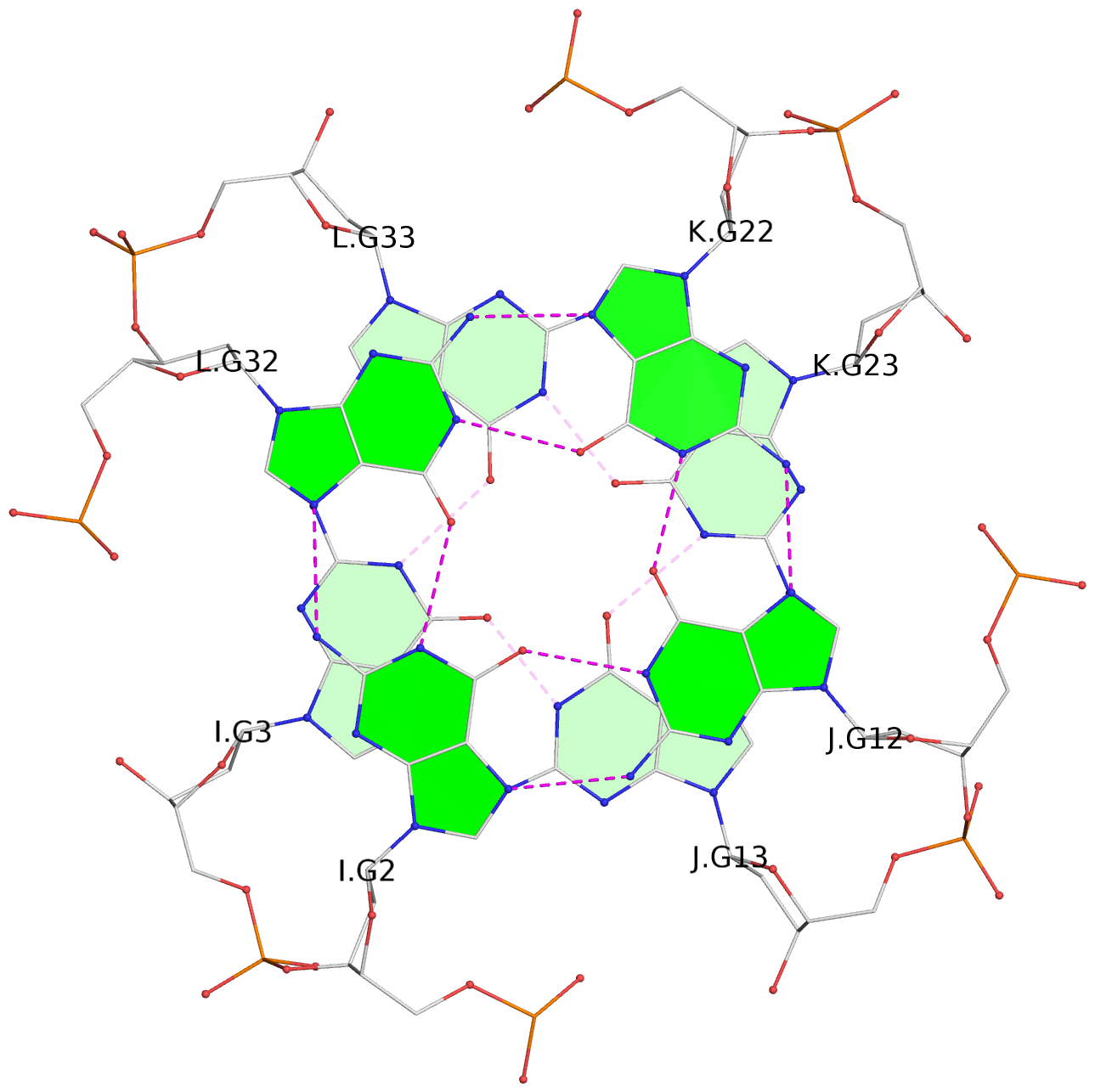
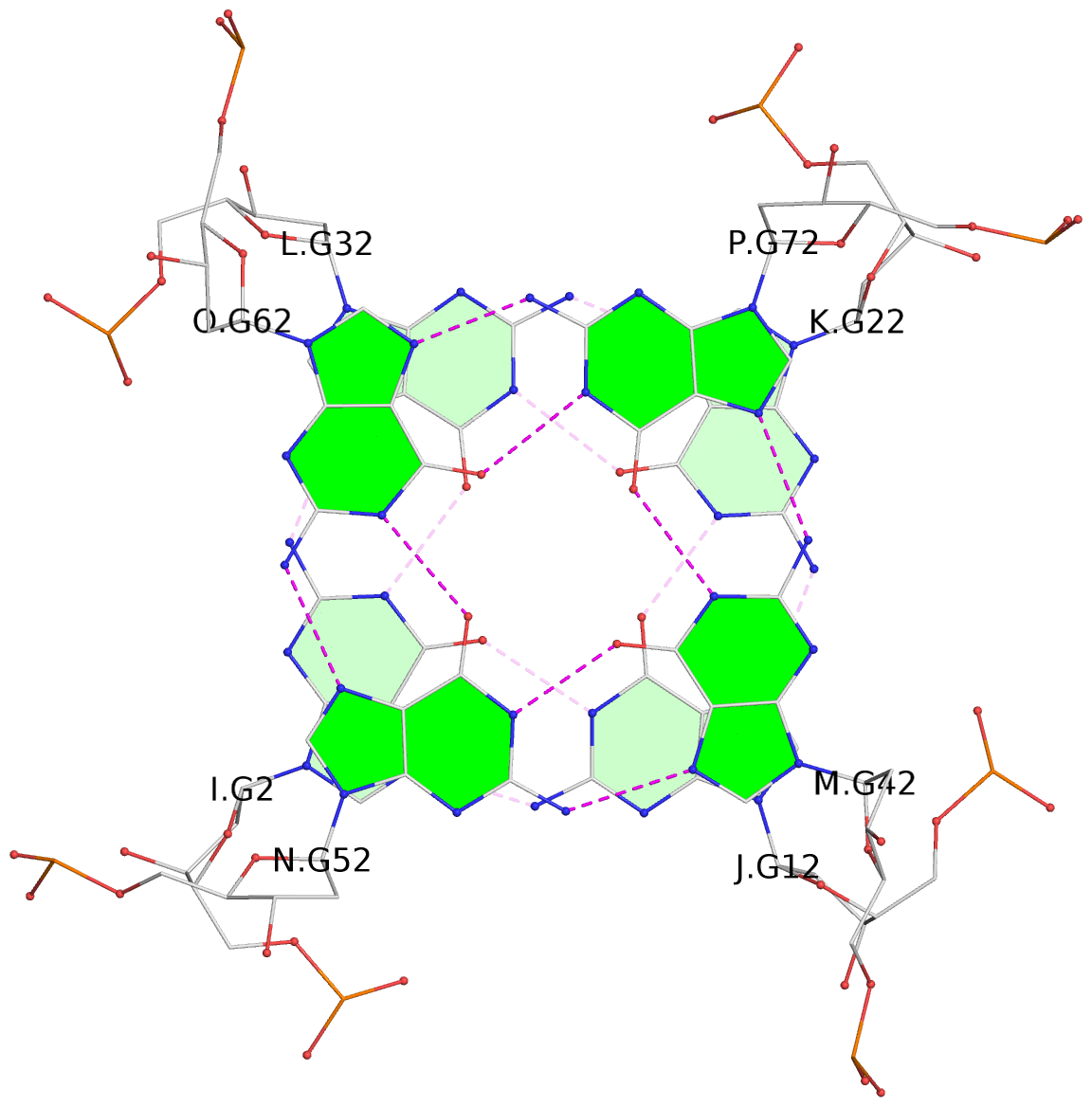
- 16 G-tetrads, 2 G4 helices, 4 G4 stems, 2 G4 coaxial stacks, parallel(4+0), UUUU, coaxial interfaces: 5'/5'
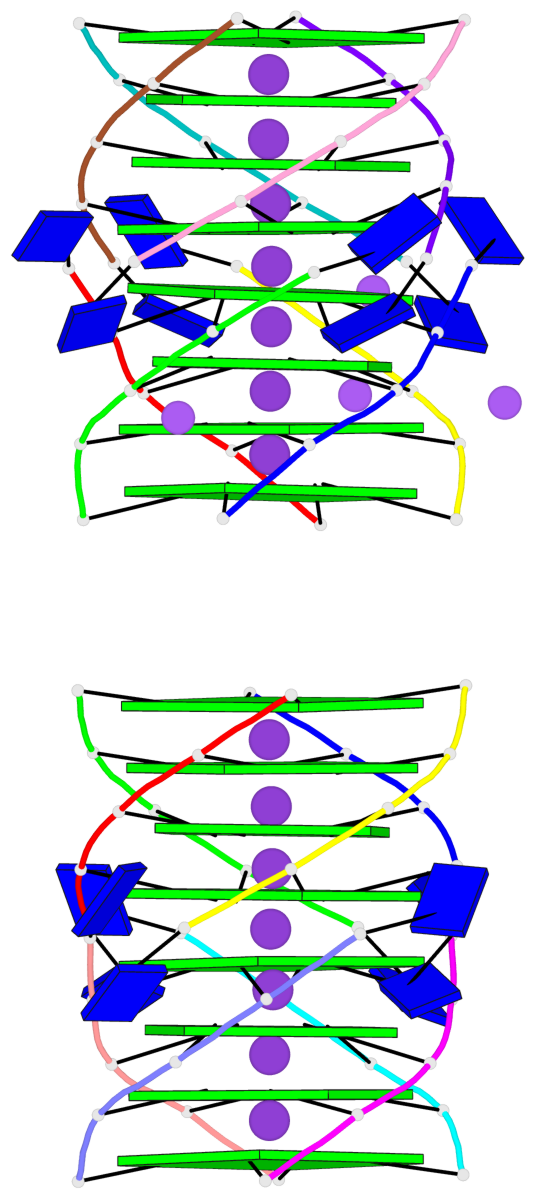
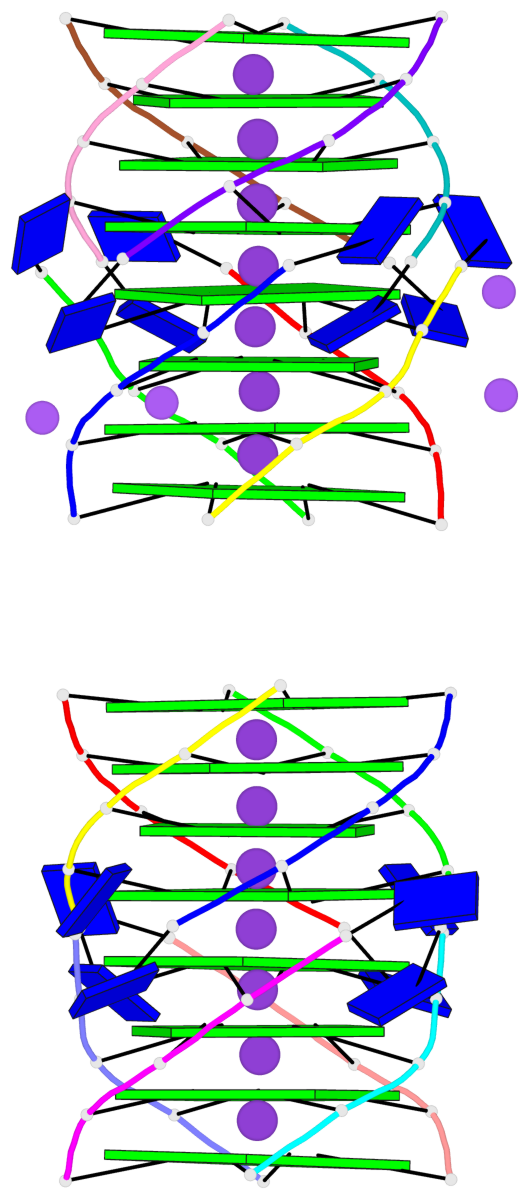
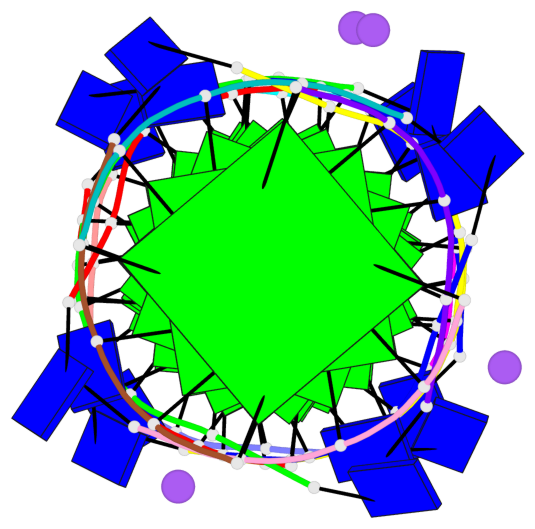
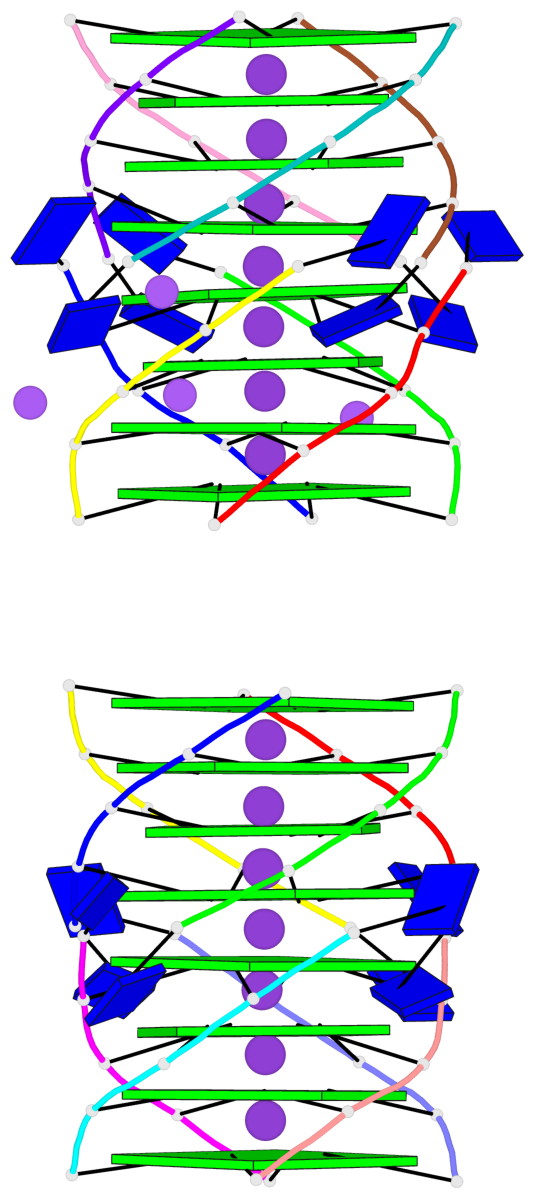
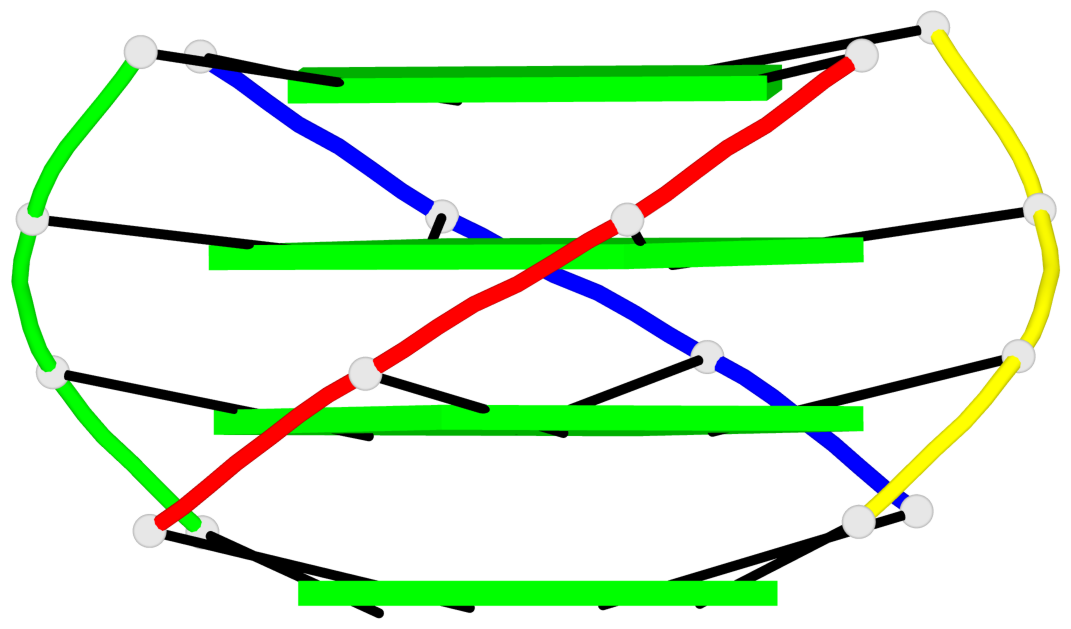
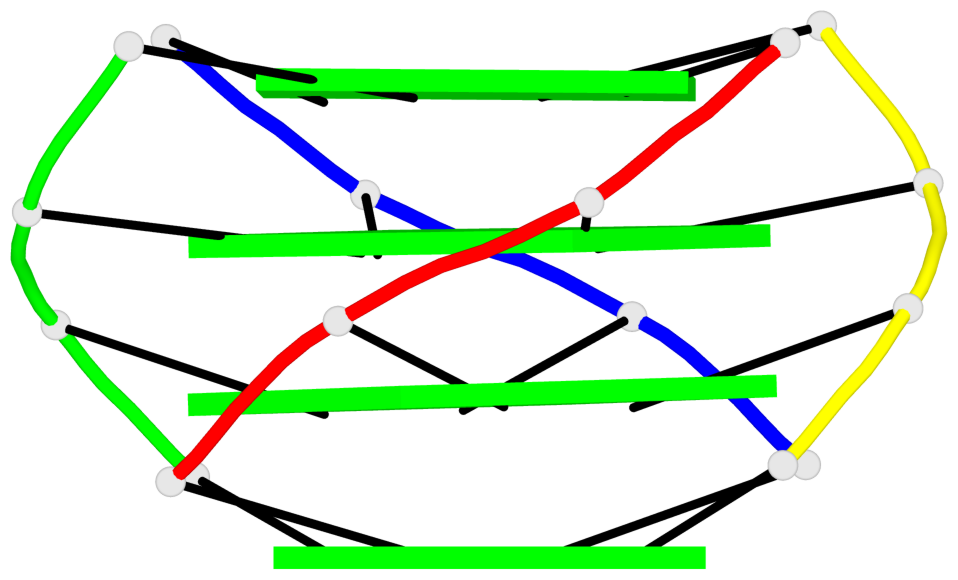
Base-block schematics in six views
List of 16 G-tetrads
1 glyco-bond=---- sugar=--.- groove=---- planarity=0.248 type=bowl nts=4 GGGG A.DG2,D.DG32,C.DG22,B.DG12 2 glyco-bond=---- sugar=---- groove=---- planarity=0.210 type=bowl nts=4 GGGG A.DG3,D.DG33,C.DG23,B.DG13 3 glyco-bond=---- sugar=---- groove=---- planarity=0.139 type=planar nts=4 GGGG A.DG4,D.DG34,C.DG24,B.DG14 4 glyco-bond=---- sugar=---- groove=---- planarity=0.198 type=bowl nts=4 GGGG A.DG5,D.DG35,C.DG25,B.DG15 5 glyco-bond=---- sugar=3333 groove=---- planarity=0.081 type=planar nts=4 GGGG E.DG42,H.DG72,G.DG62,F.DG52 6 glyco-bond=---- sugar=---- groove=---- planarity=0.293 type=bowl nts=4 GGGG E.DG43,H.DG73,G.DG63,F.DG53 7 glyco-bond=---- sugar=---- groove=---- planarity=0.201 type=bowl nts=4 GGGG E.DG44,H.DG74,G.DG64,F.DG54 8 glyco-bond=---- sugar=---- groove=---- planarity=0.243 type=bowl nts=4 GGGG E.DG45,H.DG75,G.DG65,F.DG55 9 glyco-bond=---- sugar=-3-- groove=---- planarity=0.208 type=bowl nts=4 GGGG I.DG2,L.DG32,K.DG22,J.DG12 10 glyco-bond=---- sugar=---- groove=---- planarity=0.187 type=other nts=4 GGGG I.DG3,L.DG33,K.DG23,J.DG13 11 glyco-bond=---- sugar=---- groove=---- planarity=0.155 type=planar nts=4 GGGG I.DG4,L.DG34,K.DG24,J.DG14 12 glyco-bond=---- sugar=---- groove=---- planarity=0.204 type=bowl nts=4 GGGG I.DG5,L.DG35,K.DG25,J.DG15 13 glyco-bond=---- sugar=3333 groove=---- planarity=0.071 type=planar nts=4 GGGG M.DG42,P.DG72,O.DG62,N.DG52 14 glyco-bond=---- sugar=---- groove=---- planarity=0.218 type=bowl nts=4 GGGG M.DG43,P.DG73,O.DG63,N.DG53 15 glyco-bond=---- sugar=3--- groove=---- planarity=0.204 type=bowl nts=4 GGGG M.DG44,P.DG74,O.DG64,N.DG54 16 glyco-bond=---- sugar=---- groove=---- planarity=0.363 type=bowl nts=4 GGGG M.DG45,P.DG75,O.DG65,N.DG55
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 8 G-tetrad layers, inter-molecular, with 2 stems
Helix#2, 8 G-tetrad layers, inter-molecular, with 2 stems
List of 4 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 4 G-tetrad layers, 0 loops, inter-molecular, UUUU, parallel, parallel(4+0)
Stem#2, 4 G-tetrad layers, 0 loops, inter-molecular, UUUU, parallel, parallel(4+0)
Stem#3, 4 G-tetrad layers, 0 loops, inter-molecular, UUUU, parallel, parallel(4+0)
Stem#4, 4 G-tetrad layers, 0 loops, inter-molecular, UUUU, parallel, parallel(4+0)
List of 2 G4 coaxial stacks
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [5'/5'] 2 G4 helix#2 contains 2 G4 stems: [#3,#4] [5'/5']