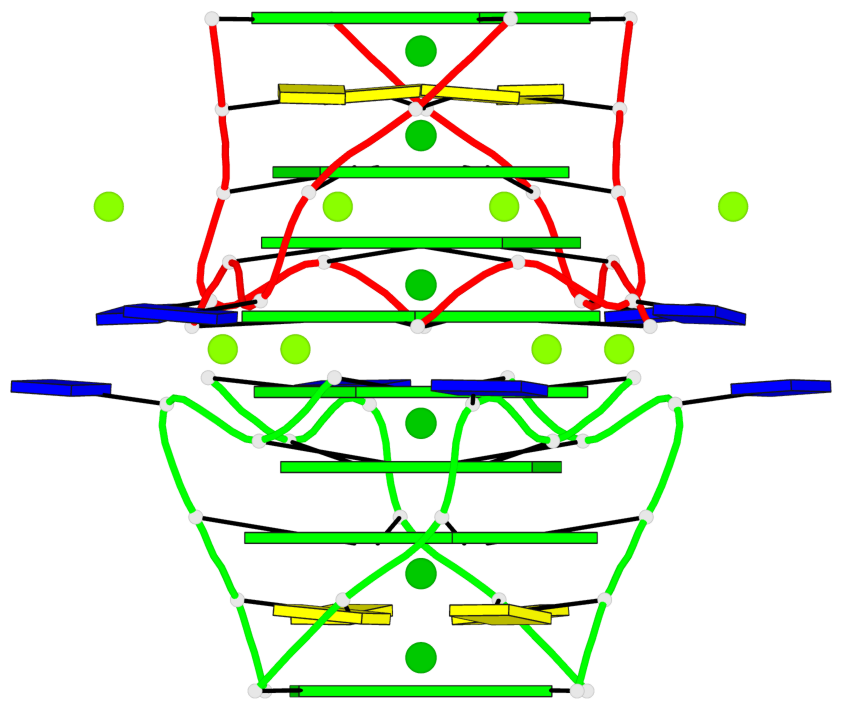
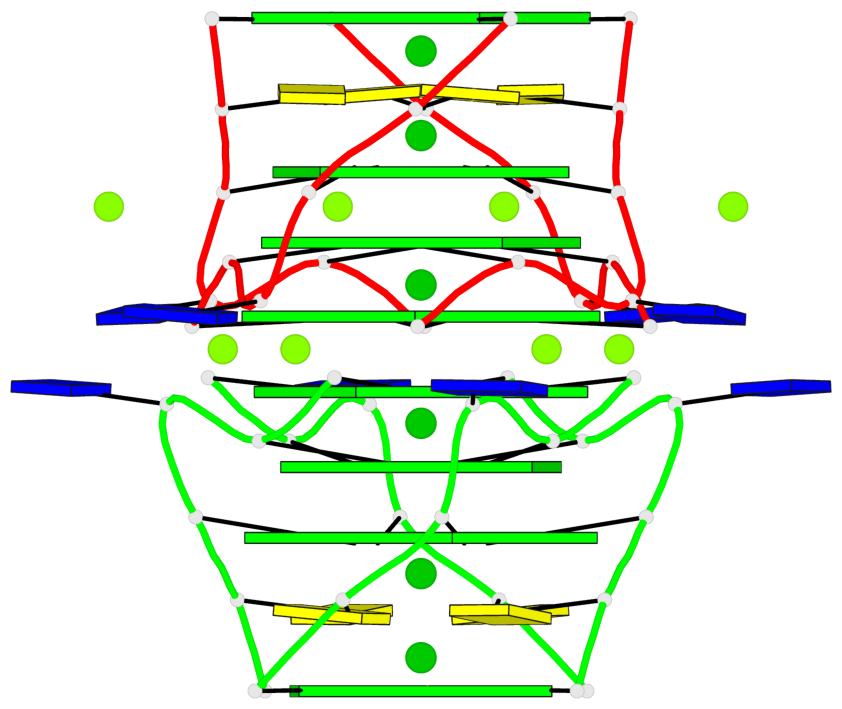
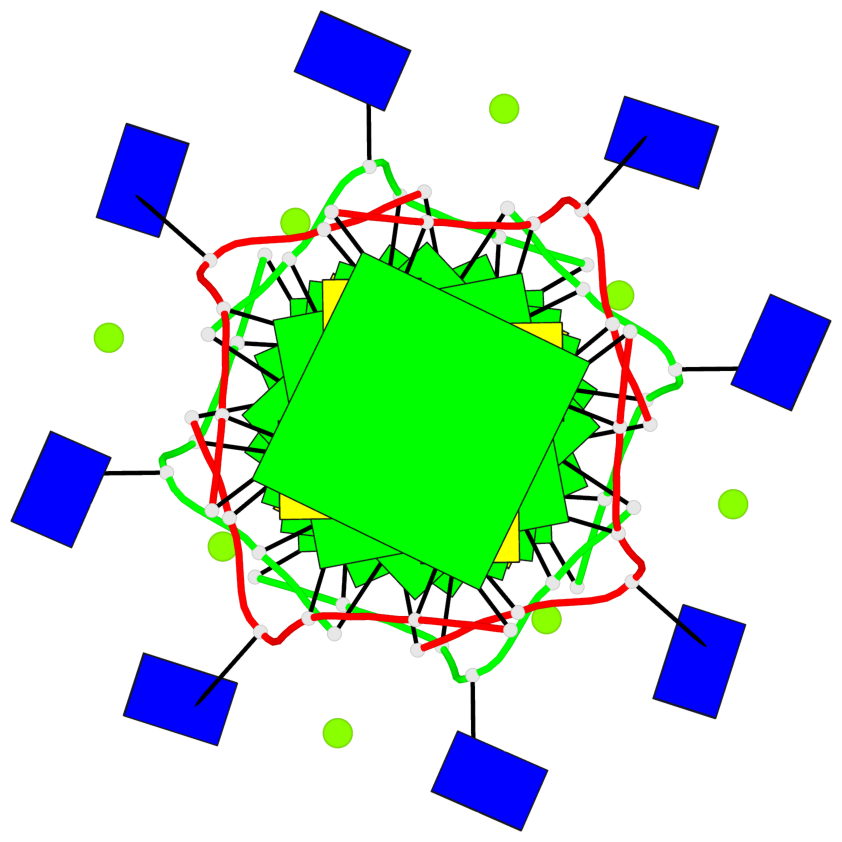
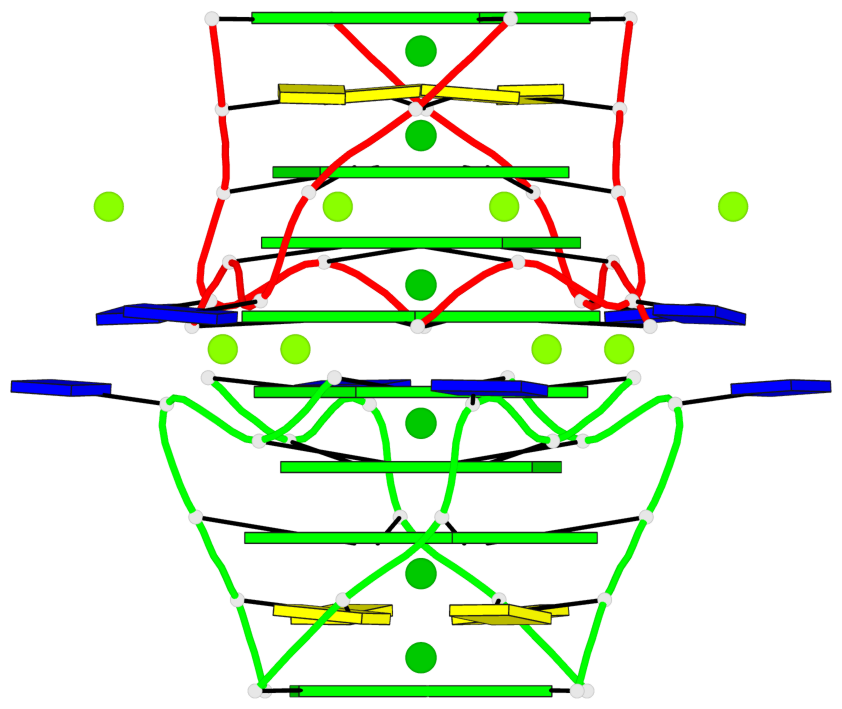
Detailed DSSR results for the G-quadruplex: PDB entry 4u92
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 4u92
- Class
- DNA
- Method
- X-ray (1.5 Å)
- Summary
- Crystal structure of a DNA-ba2+ g-quadruplex containing a water-mediated c-tetrad
- Reference
- Zhang D, Huang T, Lukeman PS, Paukstelis PJ (2014): "Crystal structure of a DNA/Ba2+ G-quadruplex containing a water-mediated C-tetrad." Nucleic Acids Res., 42, 13422-13429. doi: 10.1093/nar/gku1122.
- Abstract
- We have determined the 1.50 Å crystal structure of the DNA decamer, d(CCA(CNV)KGCGTGG) ((CNV)K, 3-cyanovinylcarbazole), which forms a G-quadruplex structure in the presence of Ba(2+). The structure contains several unique features including a bulged nucleotide and the first crystal structure observation of a C-tetrad. The structure reveals that water molecules mediate contacts between the divalent cations and the C-tetrad, allowing Ba(2+) ions to occupy adjacent steps in the central ion channel. One ordered Mg(2+) facilitates 3'-3' stacking of two quadruplexes in the asymmetric unit, while the bulged nucleotide mediates crystal contacts. Despite the high diffraction limit, the first four nucleotides including the (CNV)K nucleoside are disordered though they are still involved in crystal packing. This work suggests that the bulky hydrophobic groups may locally influence the formation of non-Watson-Crick structures from otherwise complementary sequences. These observations lead to the intriguing possibility that certain types of DNA damage may act as modulators of G-quadruplex formation.
- G4 notes
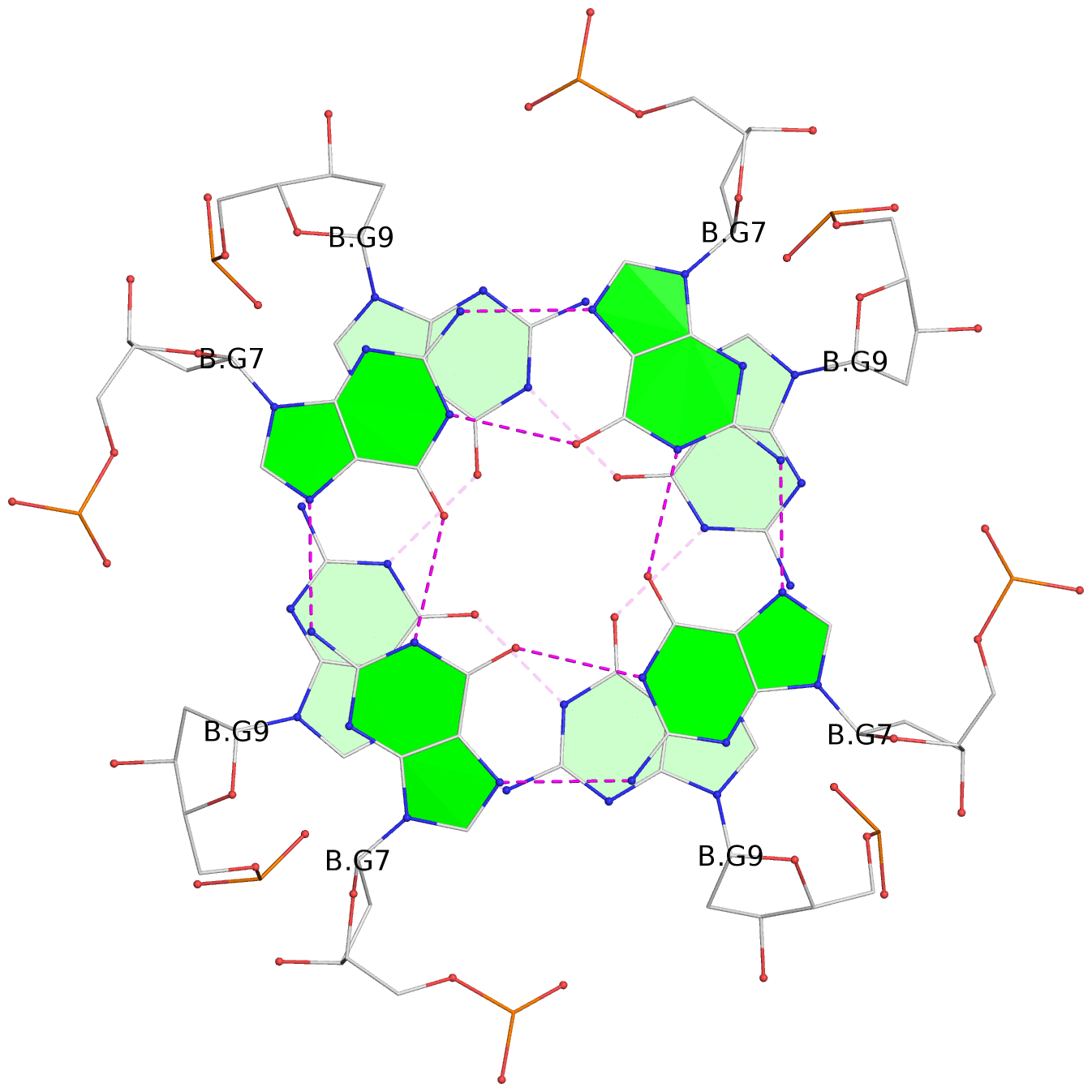
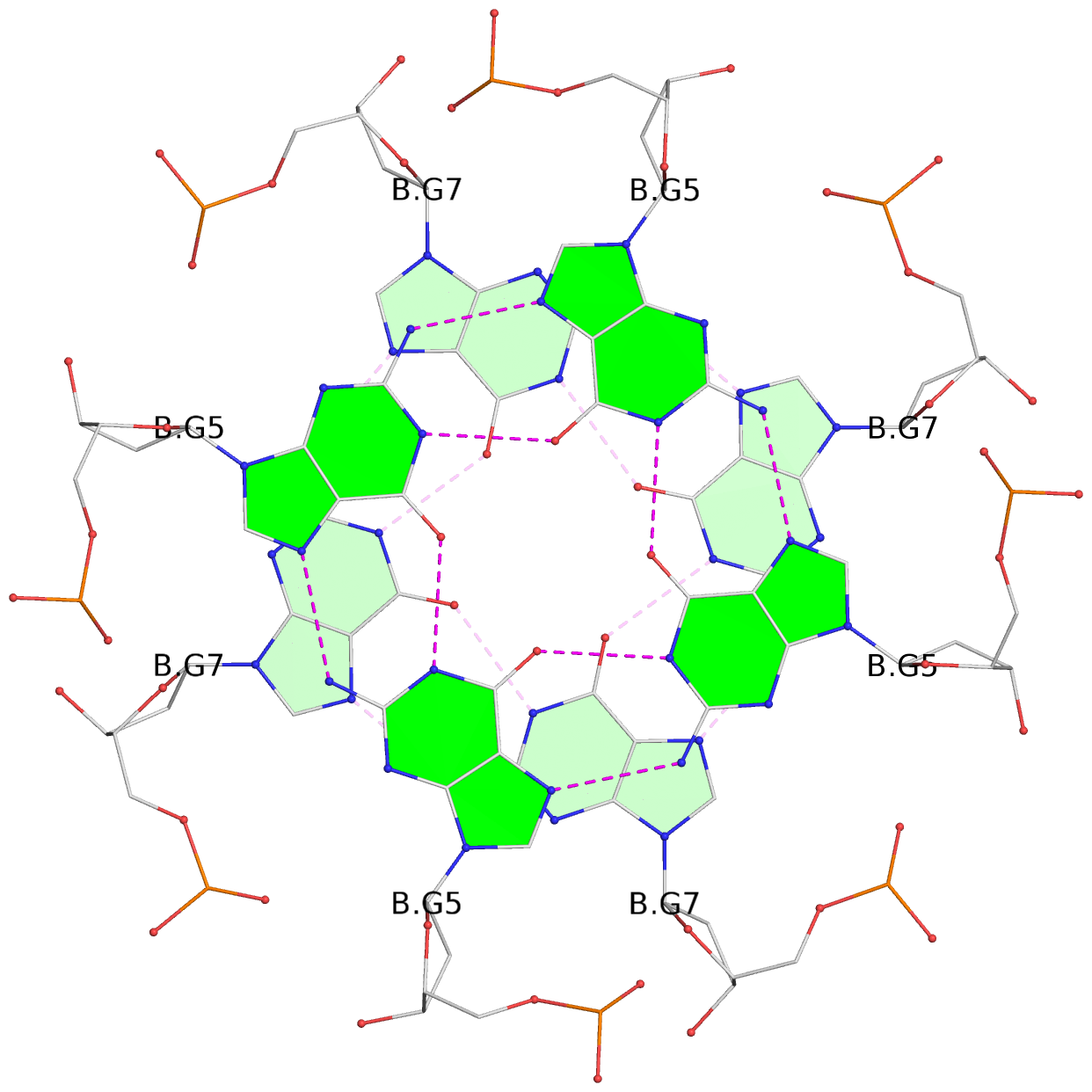
- 8 G-tetrads, 1 G4 helix, 2 G4 stems, 1 G4 coaxial stack, parallel(4+0), UUUU, coaxial interfaces: 3'/3'
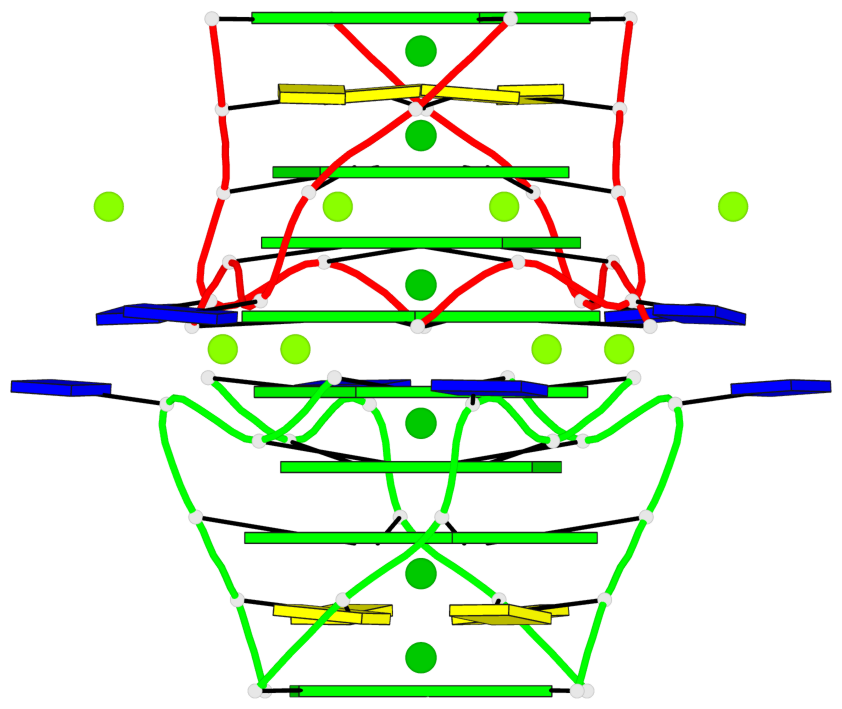
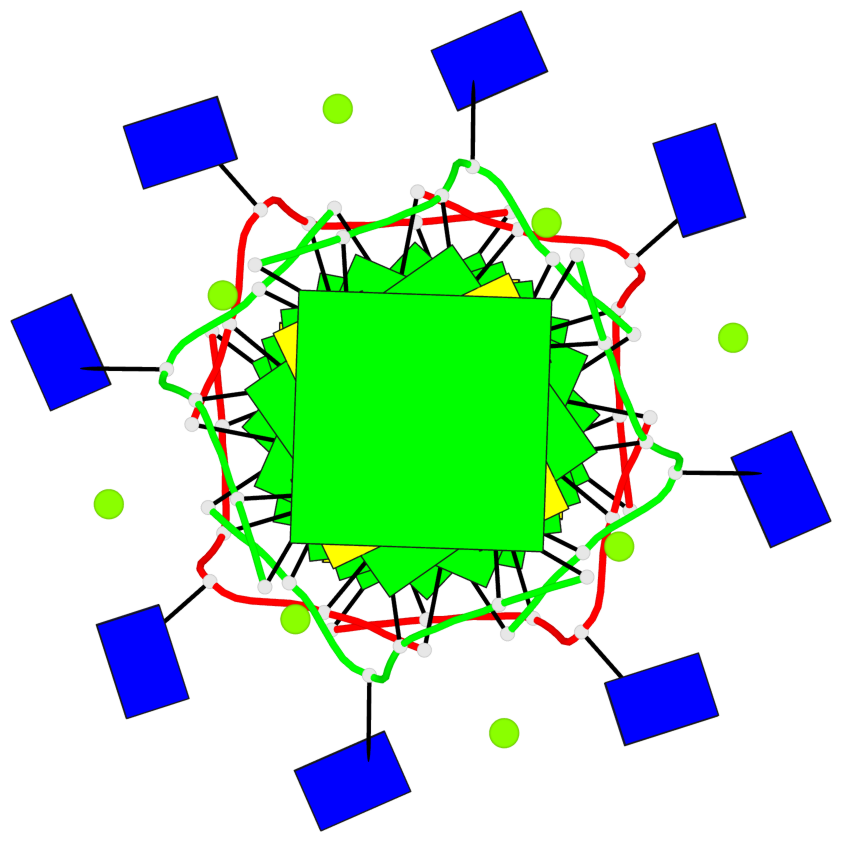
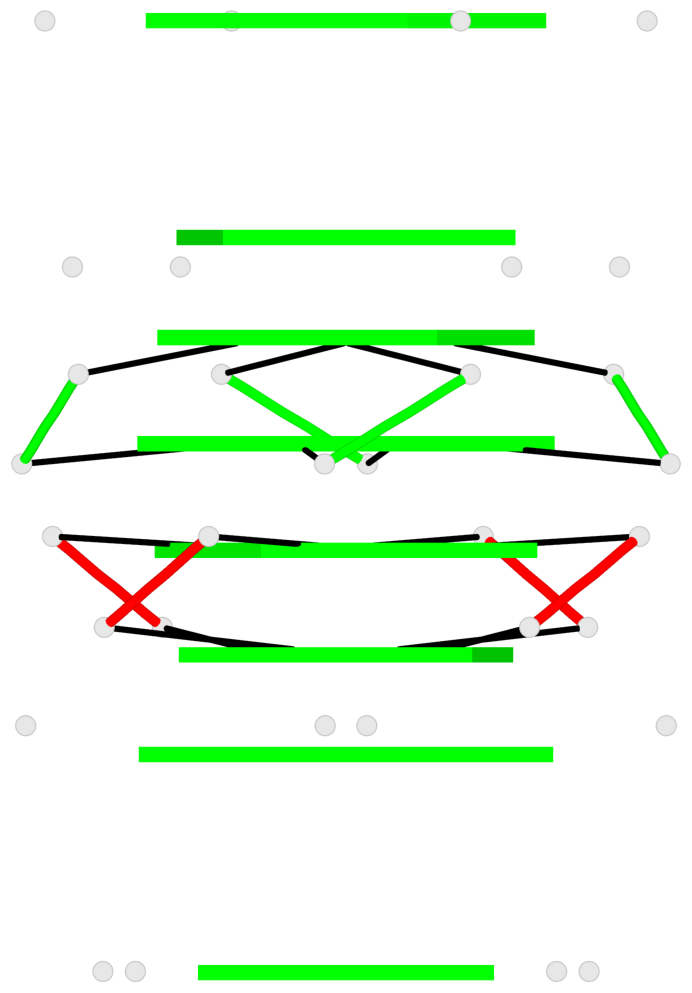
Base-block schematics in six views
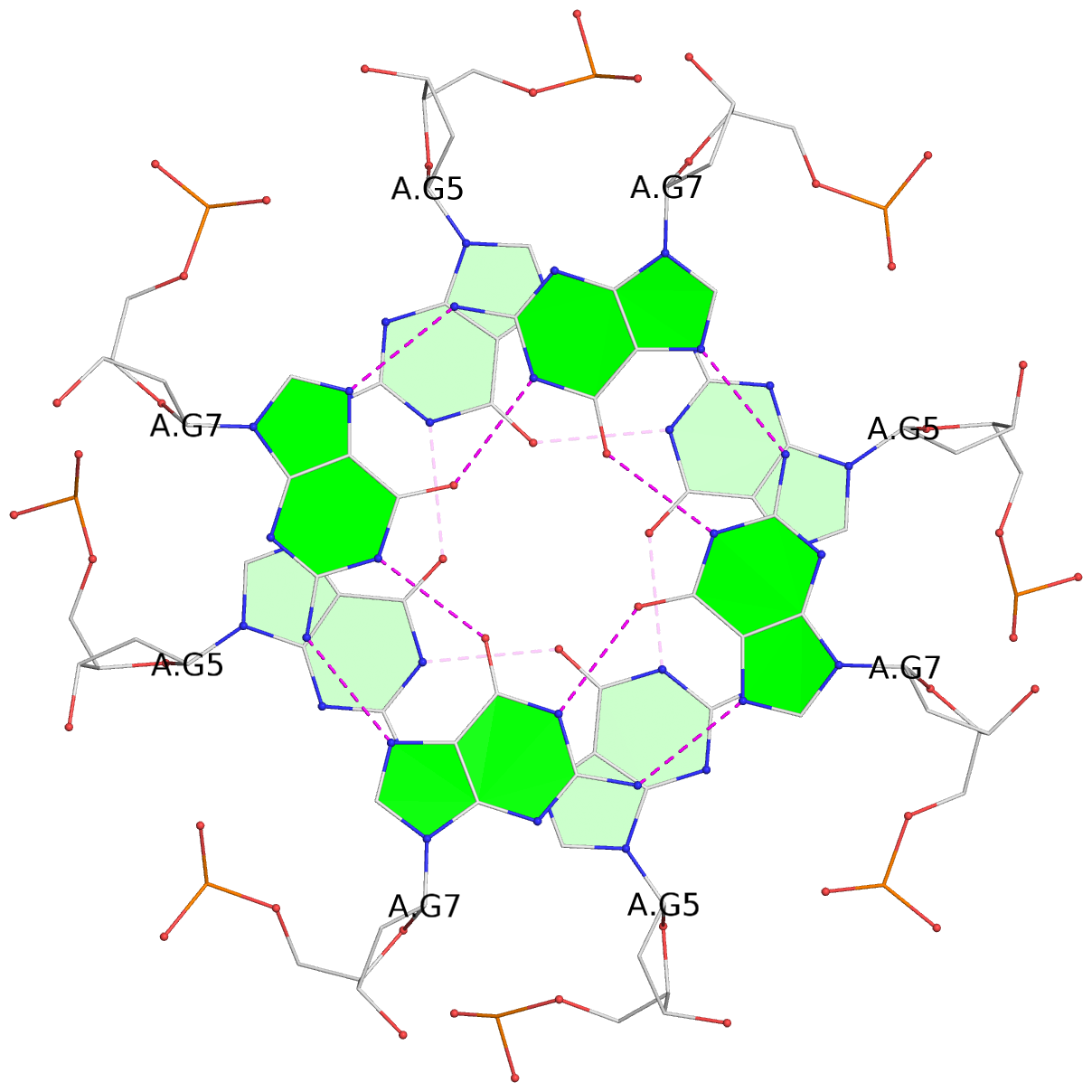
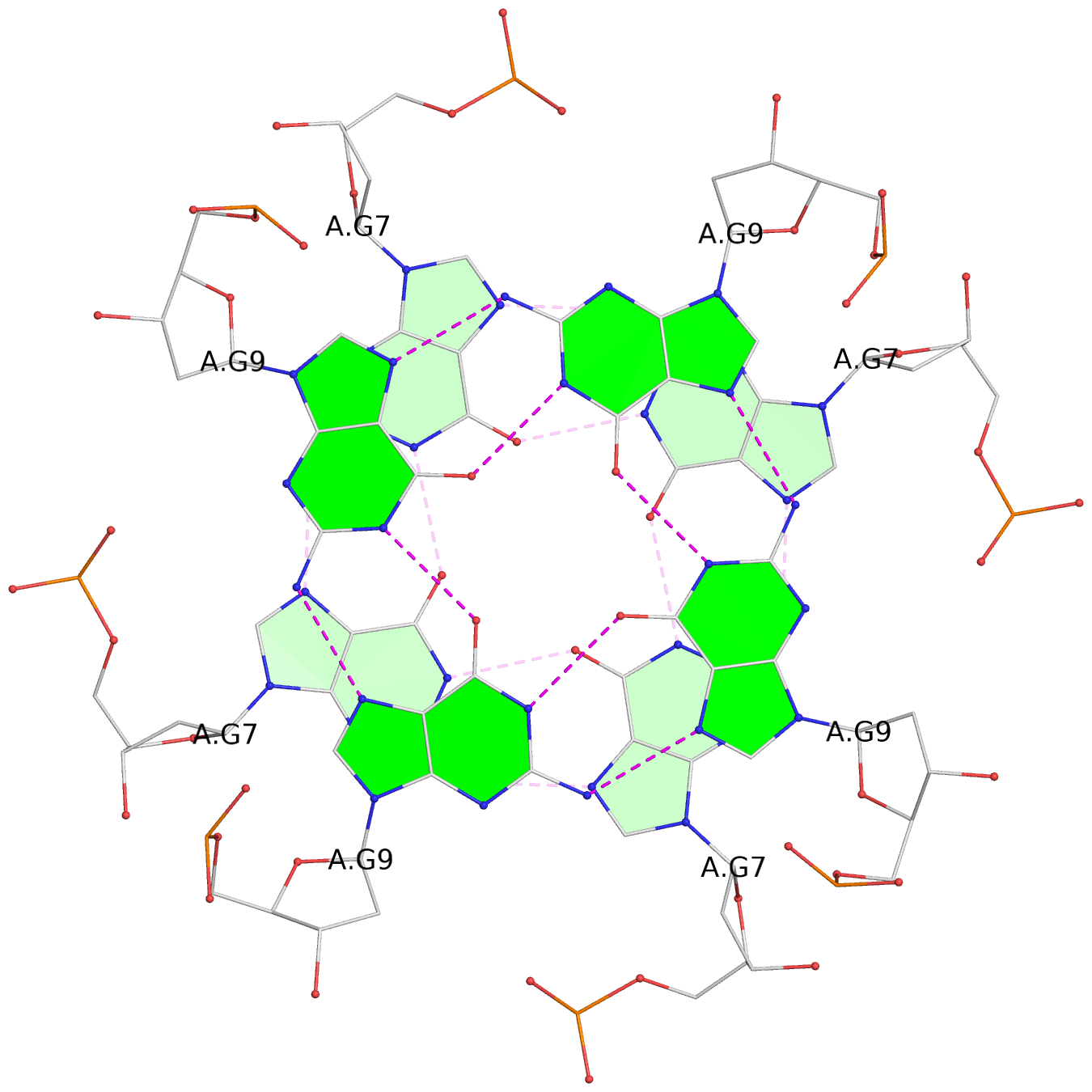
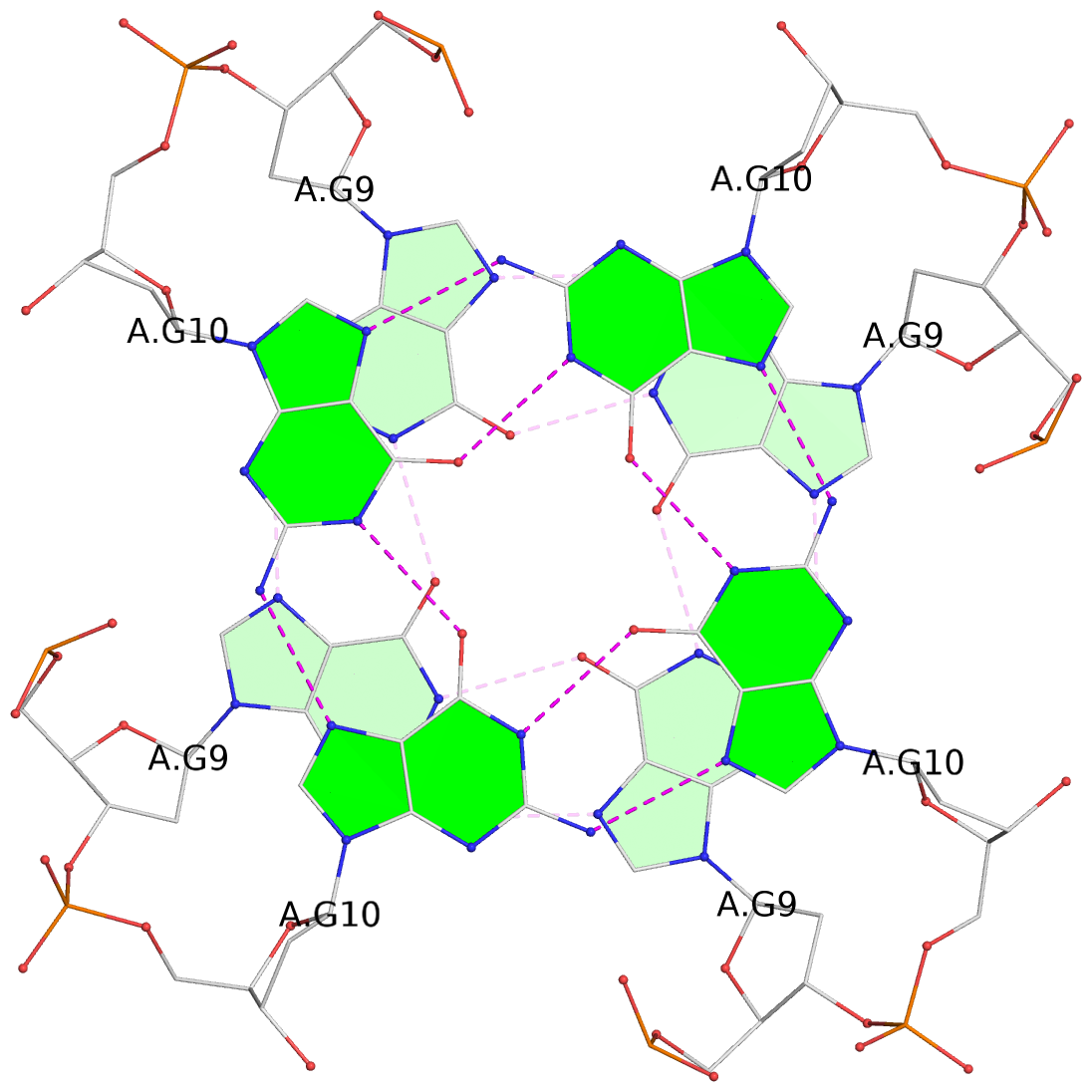
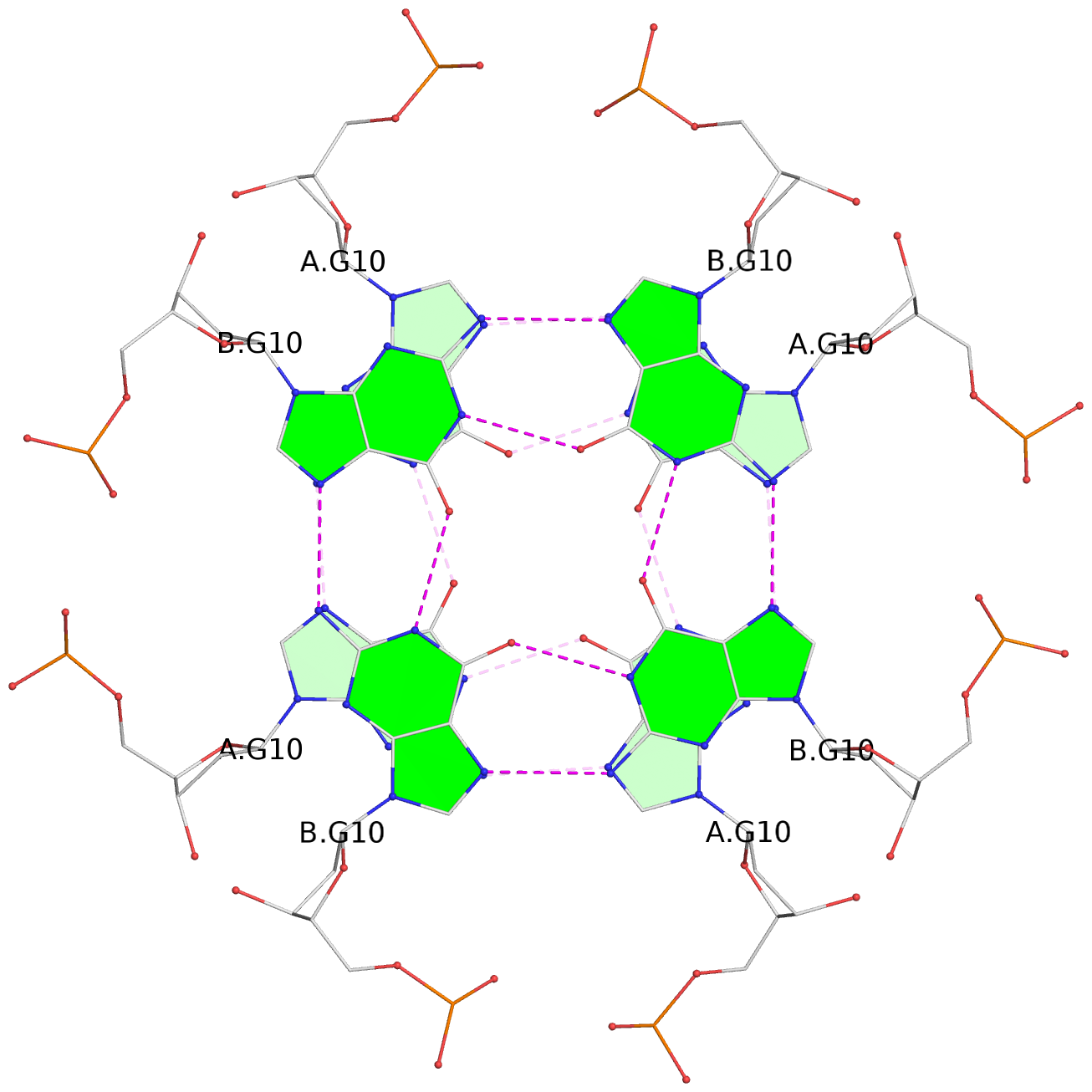
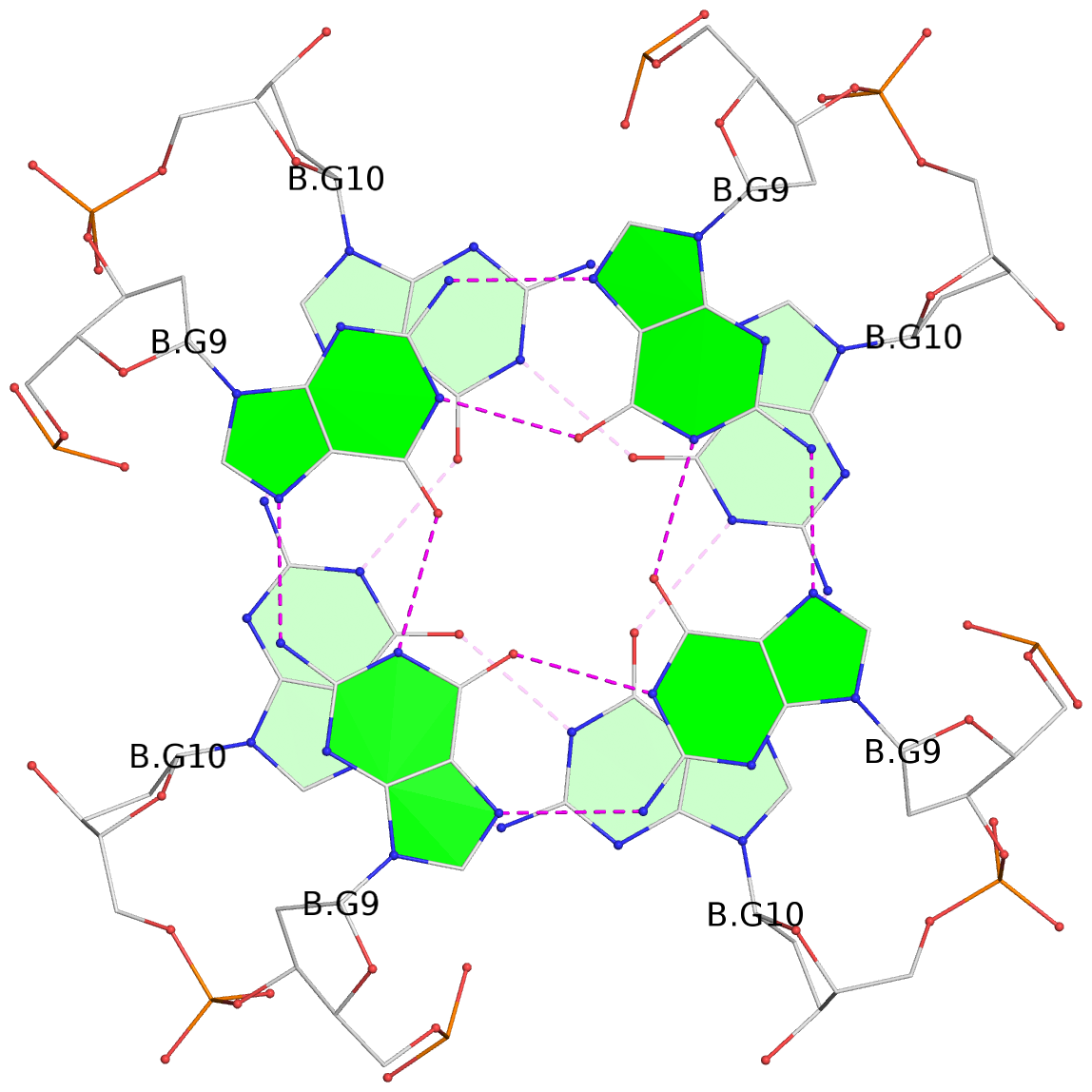
List of 8 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.105 type=planar nts=4 GGGG 1:A.DG5,4:A.DG5,2:A.DG5,3:A.DG5 2 glyco-bond=---- sugar=---- groove=---- planarity=0.216 type=bowl nts=4 GGGG 1:A.DG7,4:A.DG7,2:A.DG7,3:A.DG7 3 glyco-bond=---- sugar=3333 groove=---- planarity=0.256 type=other nts=4 GGGG 1:A.DG9,4:A.DG9,2:A.DG9,3:A.DG9 4 glyco-bond=---- sugar=---- groove=---- planarity=0.065 type=planar nts=4 GGGG 1:A.DG10,4:A.DG10,2:A.DG10,3:A.DG10 5 glyco-bond=---- sugar=---- groove=---- planarity=0.085 type=planar nts=4 GGGG 1:B.DG5,3:B.DG5,2:B.DG5,4:B.DG5 6 glyco-bond=---- sugar=---- groove=---- planarity=0.217 type=bowl nts=4 GGGG 1:B.DG7,3:B.DG7,2:B.DG7,4:B.DG7 7 glyco-bond=---- sugar=3333 groove=---- planarity=0.244 type=other nts=4 GGGG 1:B.DG9,3:B.DG9,2:B.DG9,4:B.DG9 8 glyco-bond=---- sugar=---- groove=---- planarity=0.046 type=planar nts=4 GGGG 1:B.DG10,3:B.DG10,2:B.DG10,4:B.DG10
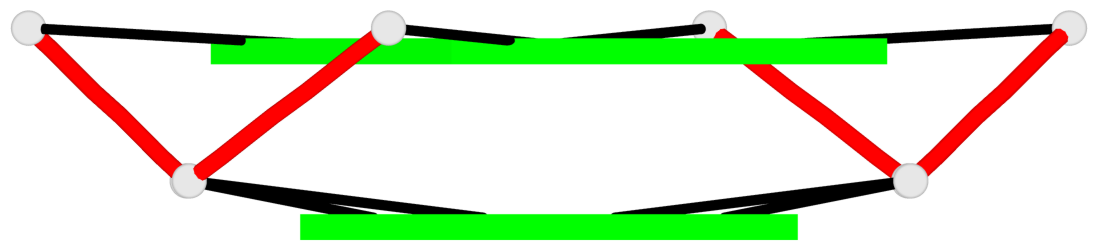
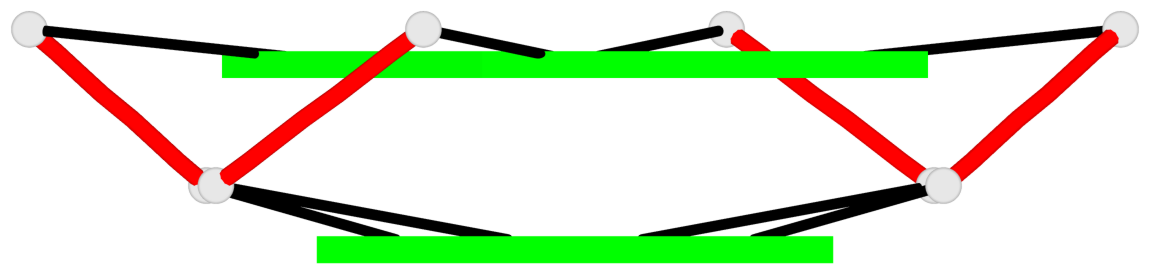
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 8 G-tetrad layers, inter-molecular, with 2 stems
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 0 loops, inter-molecular, UUUU, parallel, parallel(4+0)
Stem#2, 2 G-tetrad layers, 0 loops, inter-molecular, UUUU, parallel, parallel(4+0)
List of 1 G4 coaxial stack
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [3'/3']