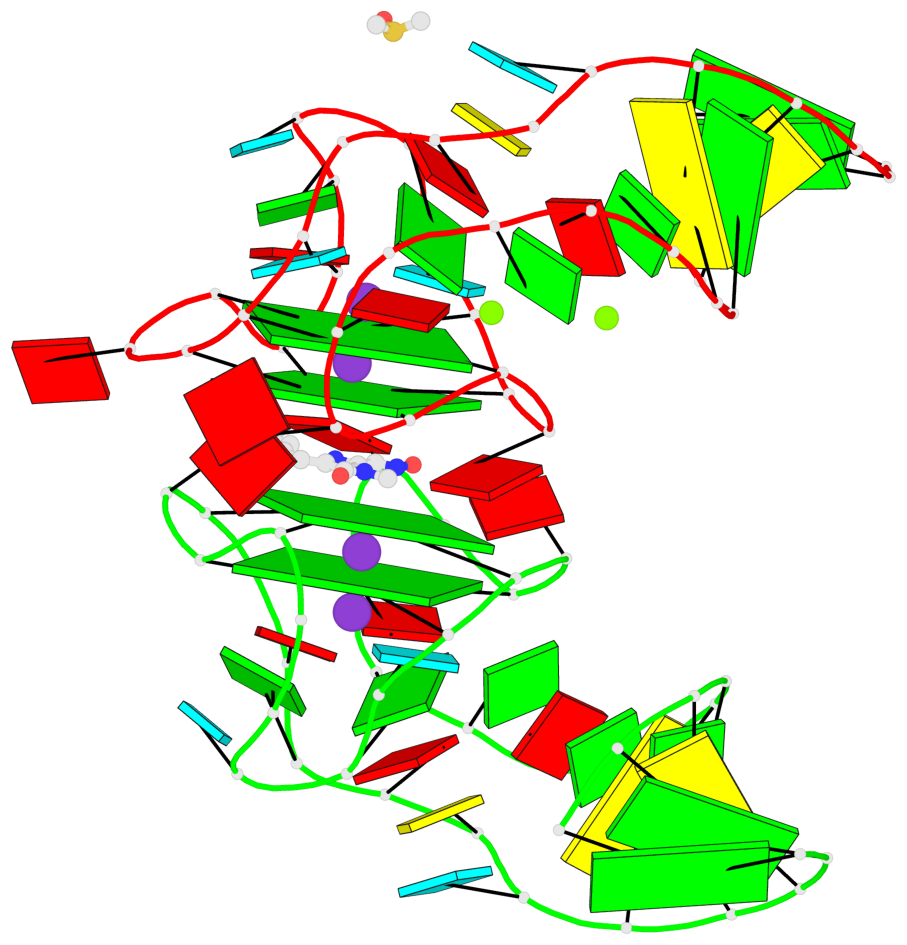
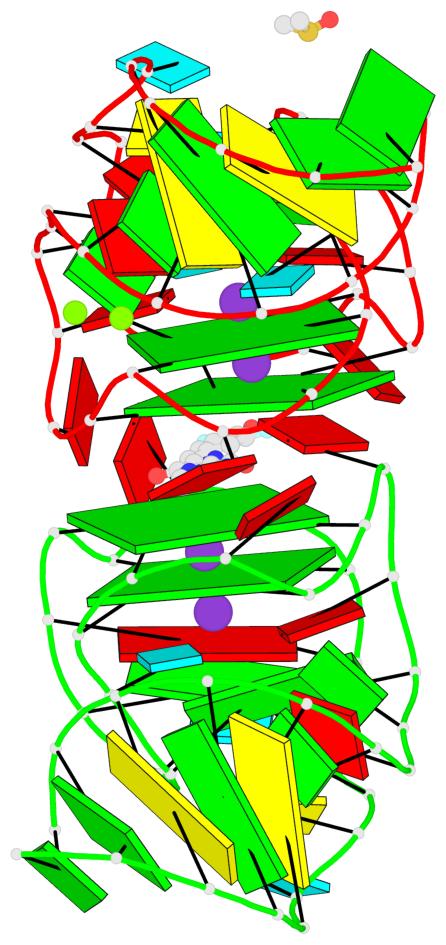
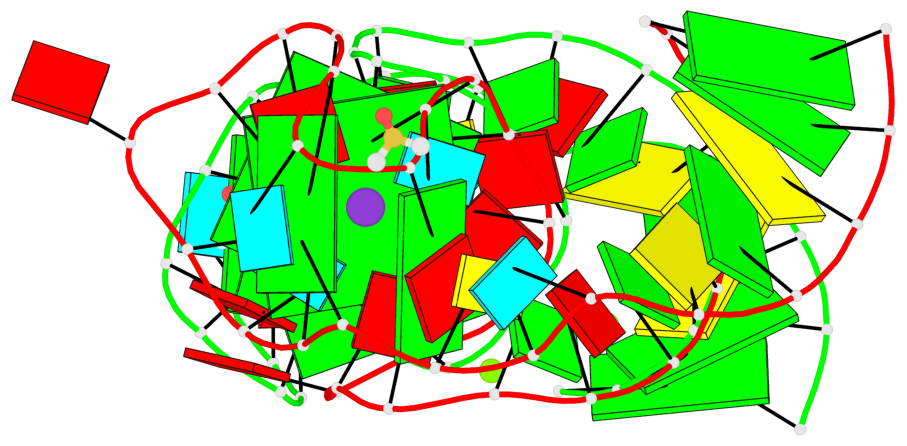
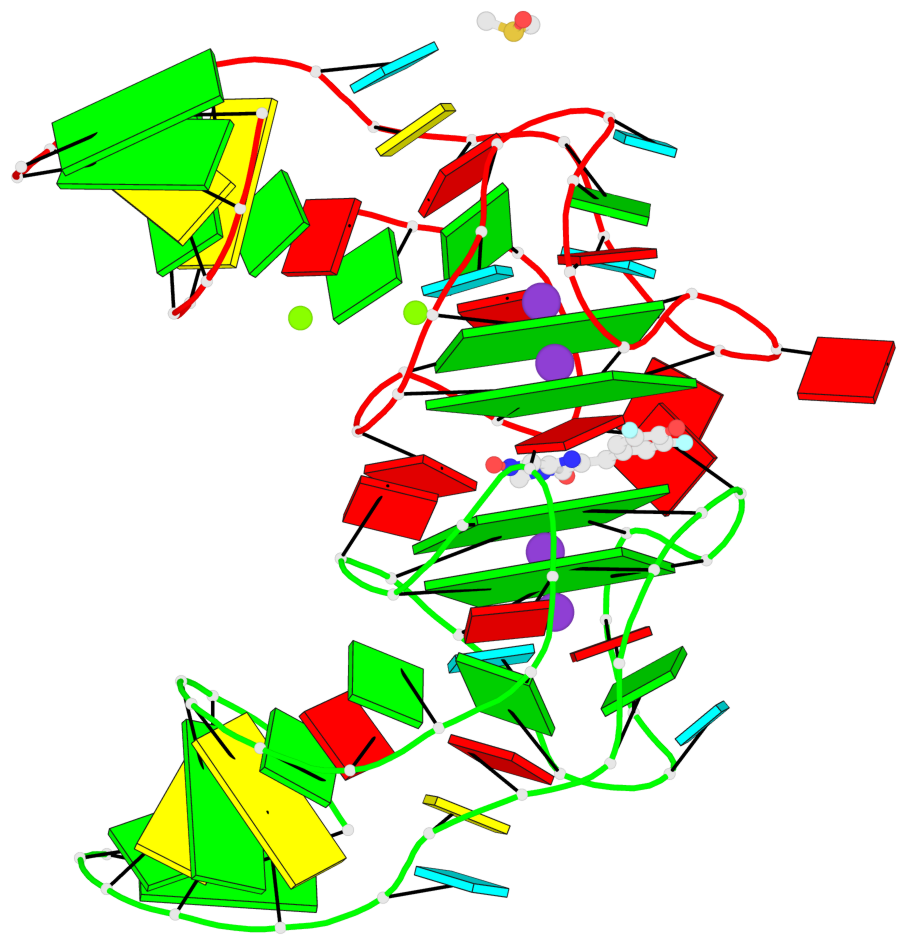
Detailed DSSR results for the G-quadruplex: PDB entry 5bjp
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 5bjp
- Class
- RNA
- Method
- X-ray (2.51 Å)
- Summary
- Crystal structure of the corn RNA aptamer in complex with dfho, iridium hexammine soak
- Reference
- Warner KD, Sjekloca L, Song W, Filonov GS, Jaffrey SR, Ferre-D'Amare AR (2017): "A homodimer interface without base pairs in an RNA mimic of red fluorescent protein." Nat. Chem. Biol., 13, 1195-1201. doi: 10.1038/nchembio.2475.
- Abstract
- Corn, a 28-nucleotide RNA, increases yellow fluorescence of its cognate ligand 3,5-difluoro-4-hydroxybenzylidene-imidazolinone-2-oxime (DFHO) by >400-fold. Corn was selected in vitro to overcome limitations of other fluorogenic RNAs, particularly rapid photobleaching. We now report the Corn-DFHO co-crystal structure, discovering that the functional species is a quasisymmetric homodimer. Unusually, the dimer interface, in which six unpaired adenosines break overall two-fold symmetry, lacks any intermolecular base pairs. The homodimer encapsulates one DFHO at its interprotomer interface, sandwiching it with a G-quadruplex from each protomer. Corn and the green-fluorescent Spinach RNA are structurally unrelated. Their convergent use of G-quadruplexes underscores the usefulness of this motif for RNA-induced small-molecule fluorescence. The asymmetric dimer interface of Corn could provide a basis for the development of mutants that only fluoresce as heterodimers. Such variants would be analogous to Split GFP, and may be useful for analyzing RNA co-expression or association, or for designing self-assembling RNA nanostructures.
- G4 notes
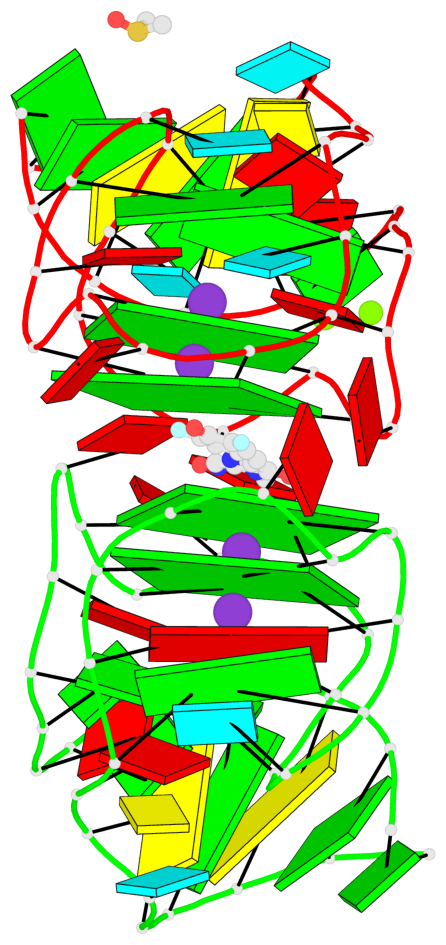
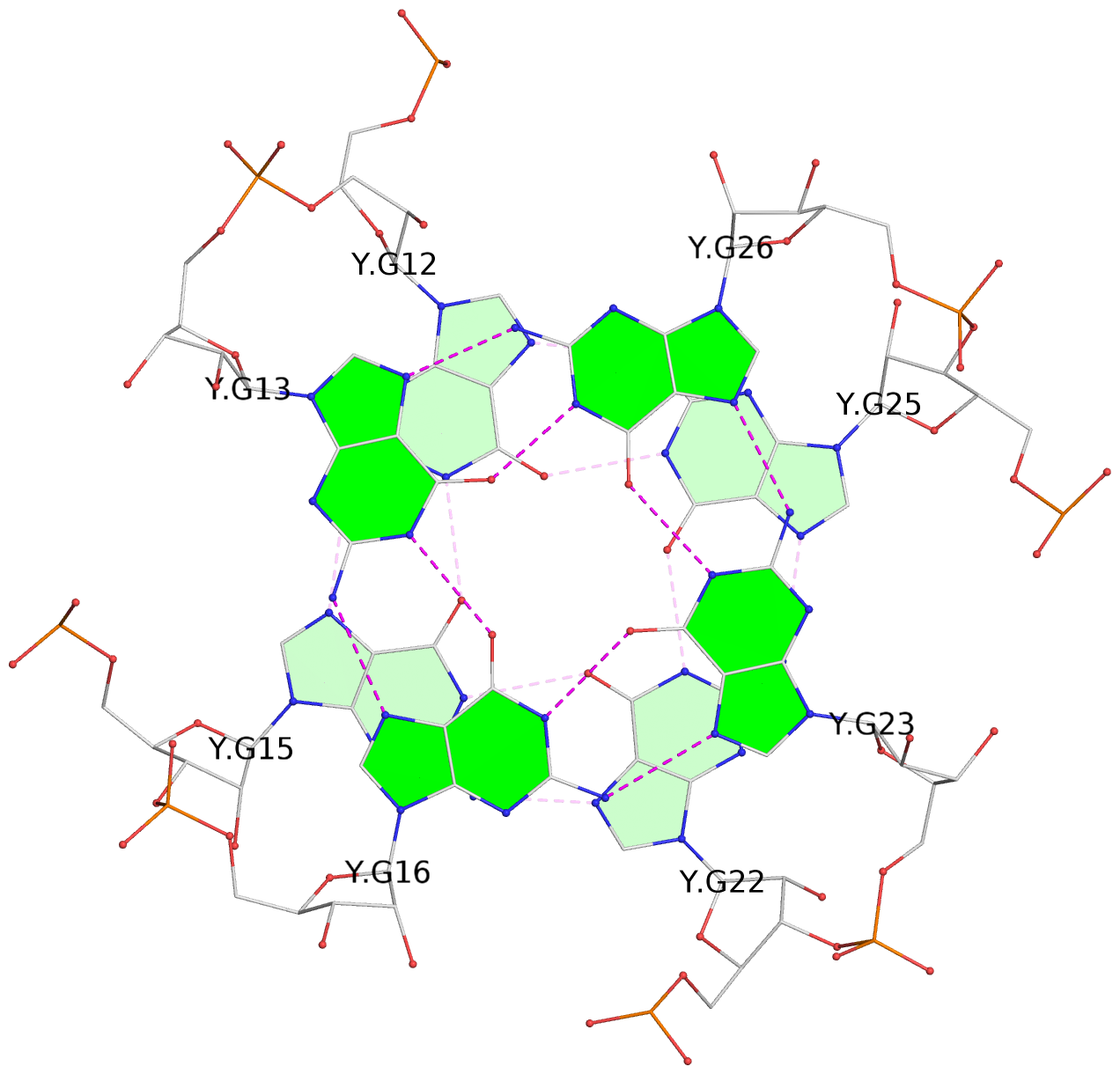
- 4 G-tetrads, 2 G4 helices, 2 G4 stems, 2(-P-P-P), parallel(4+0), UUUU
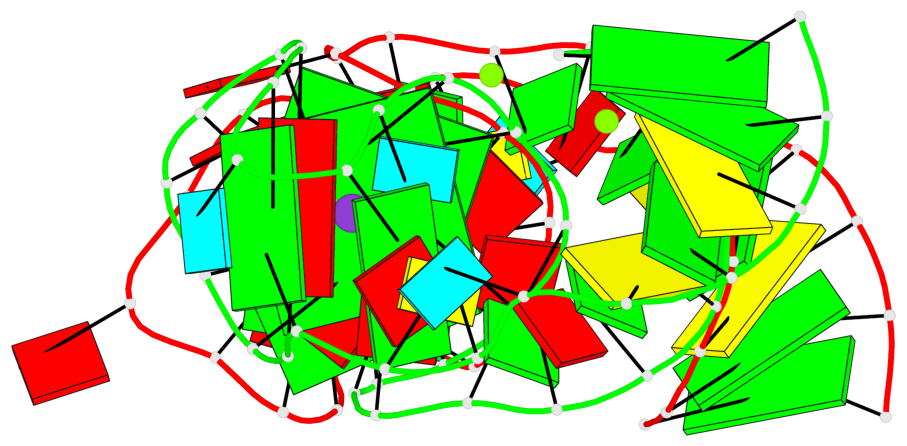
Base-block schematics in six views
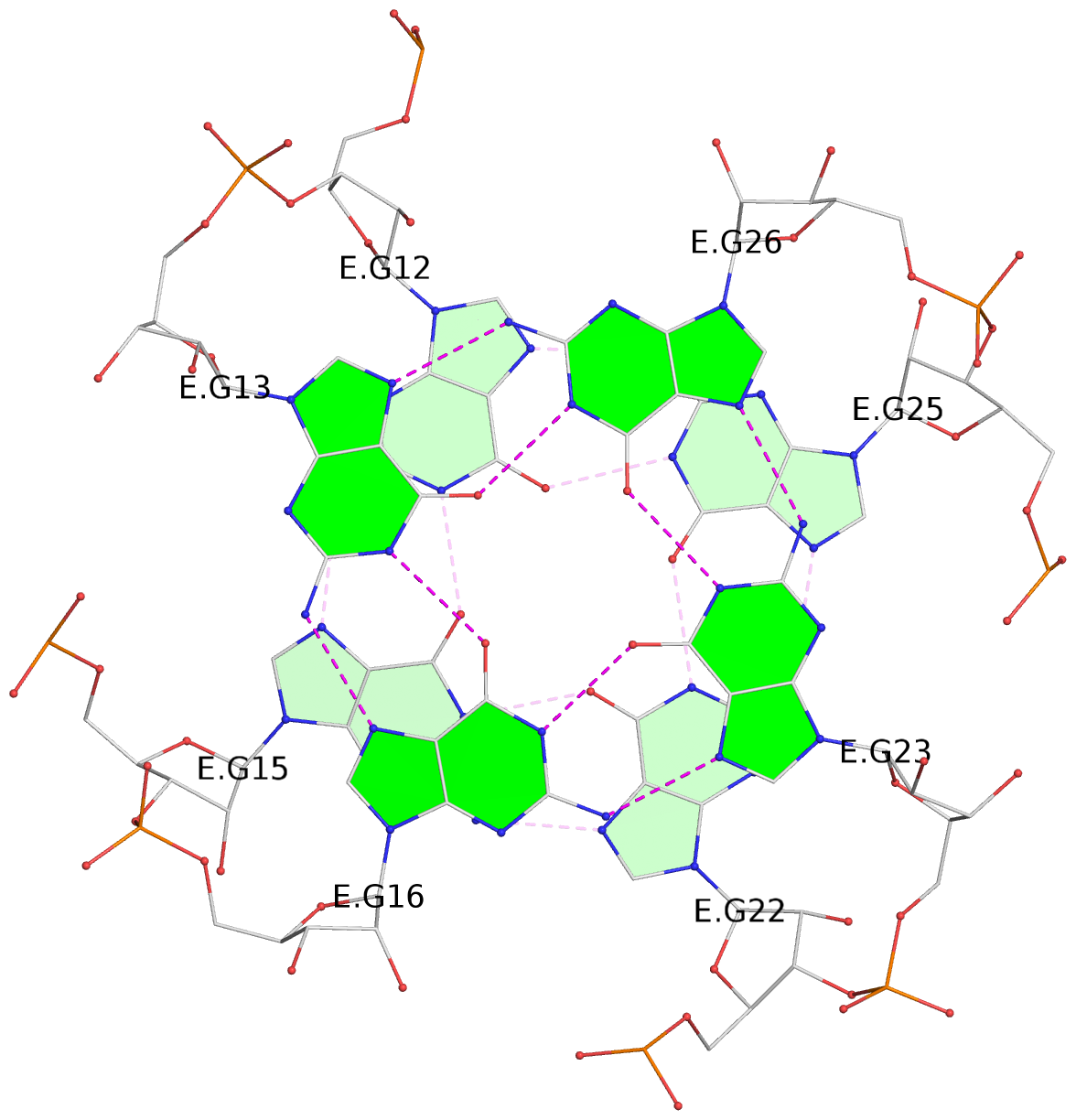
List of 4 G-tetrads
1 glyco-bond=---- sugar=-333 groove=---- planarity=0.347 type=other nts=4 GGGG E.G12,E.G15,E.G22,E.G25 2 glyco-bond=---- sugar=-3-3 groove=---- planarity=0.181 type=other nts=4 GGGG E.G13,E.G16,E.G23,E.G26 3 glyco-bond=---- sugar=-333 groove=---- planarity=0.337 type=other nts=4 GGGG Y.G12,Y.G15,Y.G22,Y.G25 4 glyco-bond=---- sugar=-3-3 groove=---- planarity=0.234 type=other nts=4 GGGG Y.G13,Y.G16,Y.G23,Y.G26
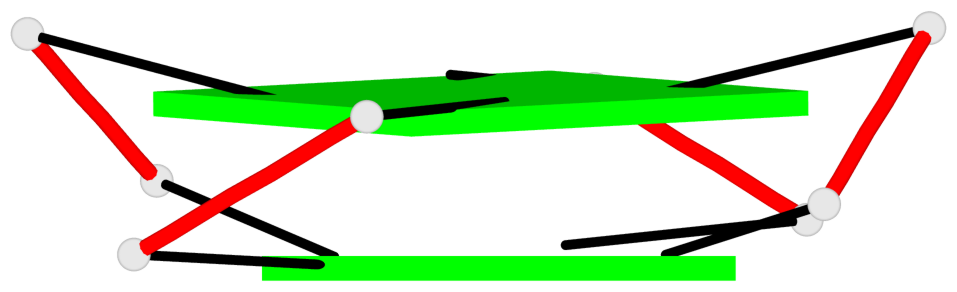
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular, with 1 stem
Helix#2, 2 G-tetrad layers, INTRA-molecular, with 1 stem
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.