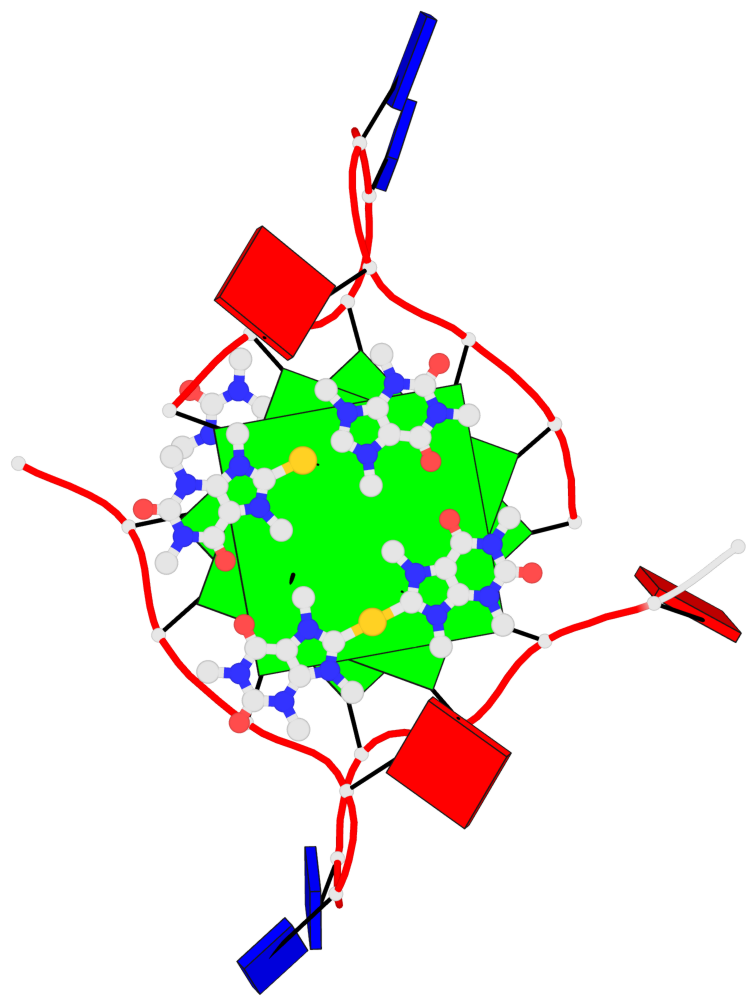
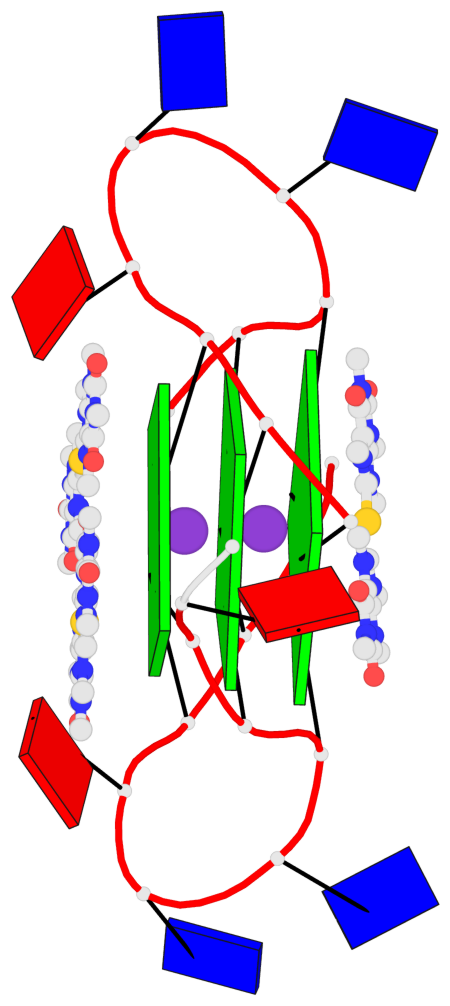
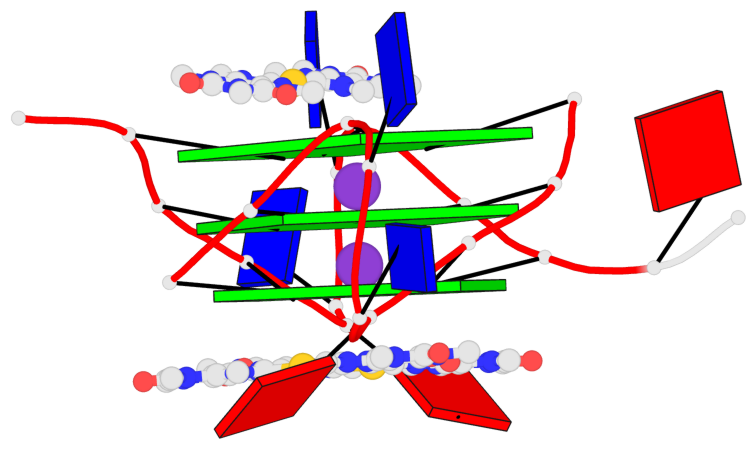
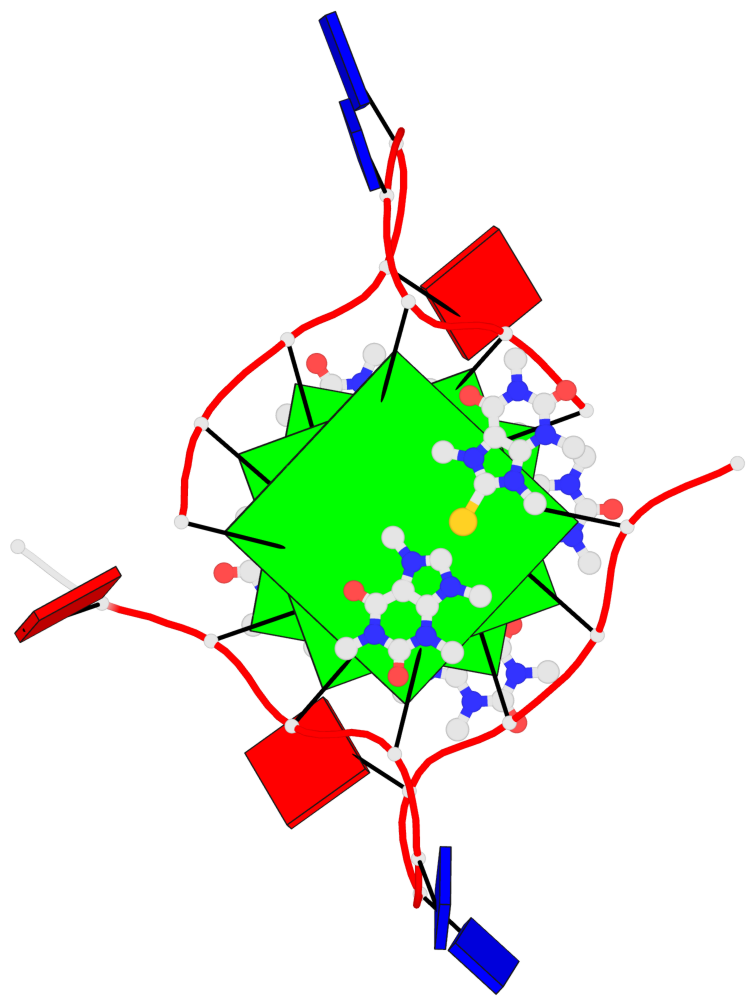
Detailed DSSR results for the G-quadruplex: PDB entry 5ccw
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 5ccw
- Class
- drug-DNA
- Method
- X-ray (1.89 Å)
- Summary
- Structure of the complex of a human telomeric DNA with au(caffein-2-ylidene)2
- Reference
- Bazzicalupi C, Ferraroni M, Papi F, Massai L, Bertrand B, Messori L, Gratteri P, Casini A (2016): "Determinants for Tight and Selective Binding of a Medicinal Dicarbene Gold(I) Complex to a Telomeric DNA G-Quadruplex: a Joint ESI MS and XRD Investigation." Angew.Chem.Int.Ed.Engl., 55, 4256-4259. doi: 10.1002/anie.201511999.
- Abstract
- The dicarbene gold(I) complex [Au(9-methylcaffein-8-ylidene)2 ]BF4 is an exceptional organometallic compound of profound interest as a prospective anticancer agent. This gold(I) complex was previously reported to be highly cytotoxic toward various cancer cell lines in vitro and behaves as a selective G-quadruplex stabilizer. Interactions of the gold complex with various telomeric DNA models have been analyzed by a combined ESI MS and X-ray diffraction (XRD) approach. ESI MS measurements confirmed formation of stable adducts between the intact gold(I) complex and Tel 23 DNA sequence. The crystal structure of the adduct formed between [Au(9-methylcaffein-8-ylidene)2 ](+) and Tel 23 DNA G-quadruplex was solved. Tel 23 maintains a characteristic propeller conformation while binding three gold(I) dicarbene moieties at two distinct sites. Stacking interactions appear to drive noncovalent binding of the gold(I) complex. The structural basis for tight gold(I) complex/G-quadruplex recognition and its selectivity are described.
- G4 notes
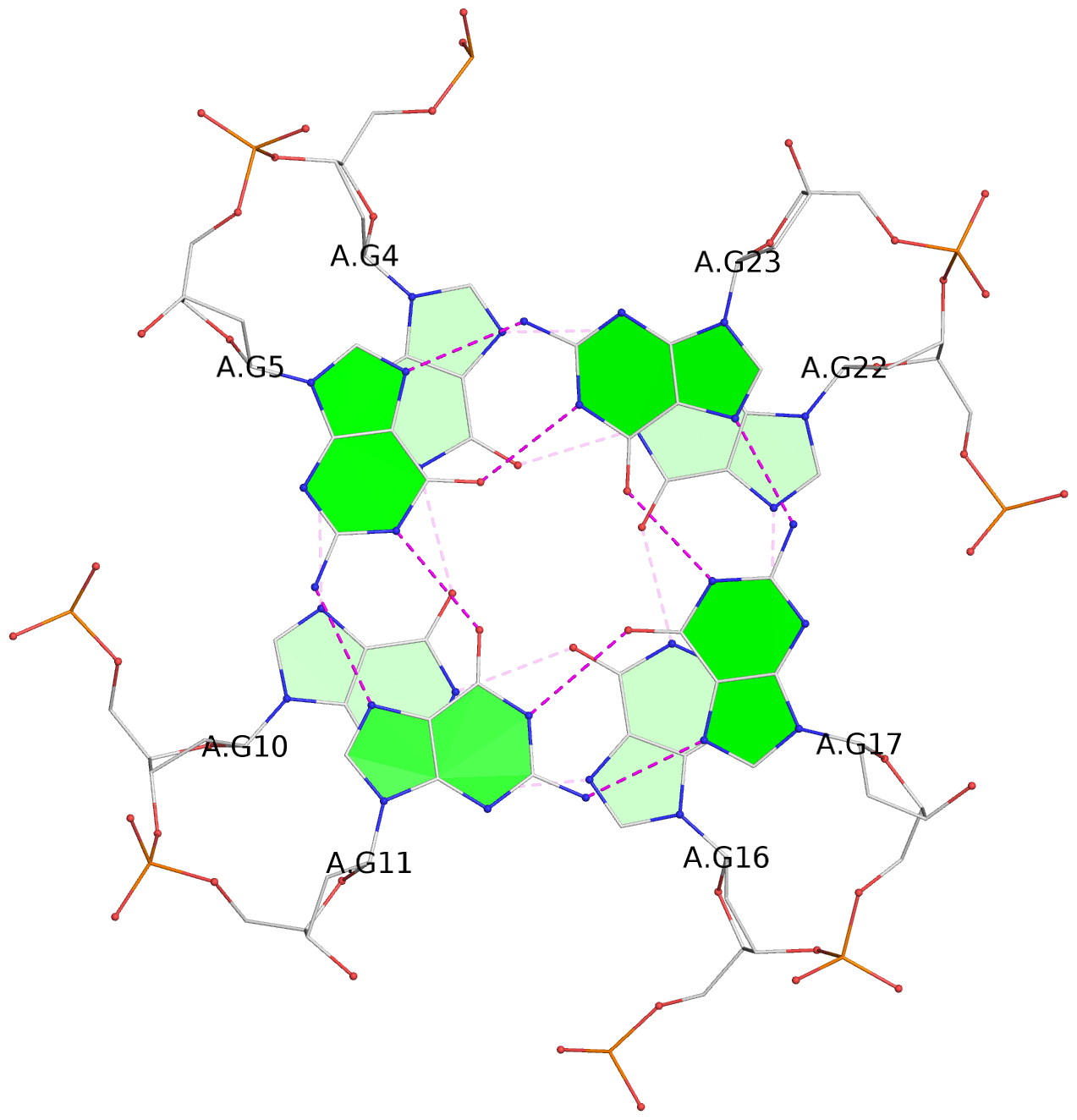
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P), parallel(4+0), UUUU
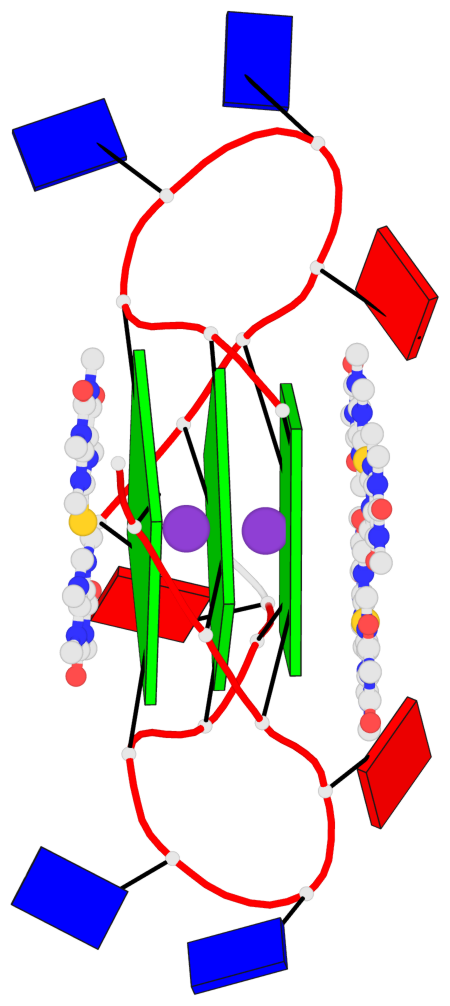
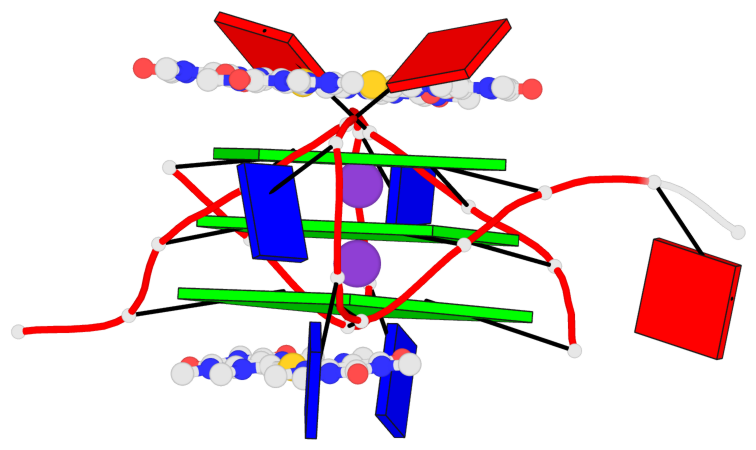
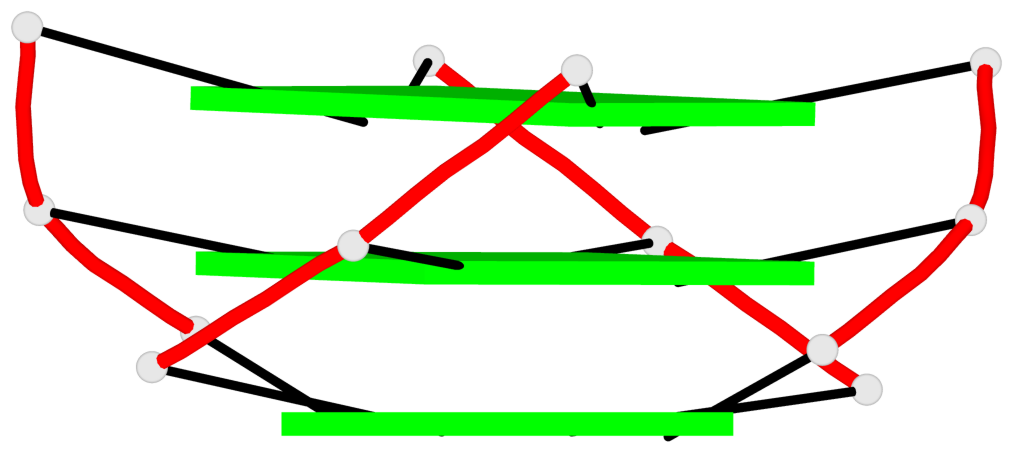
Base-block schematics in six views
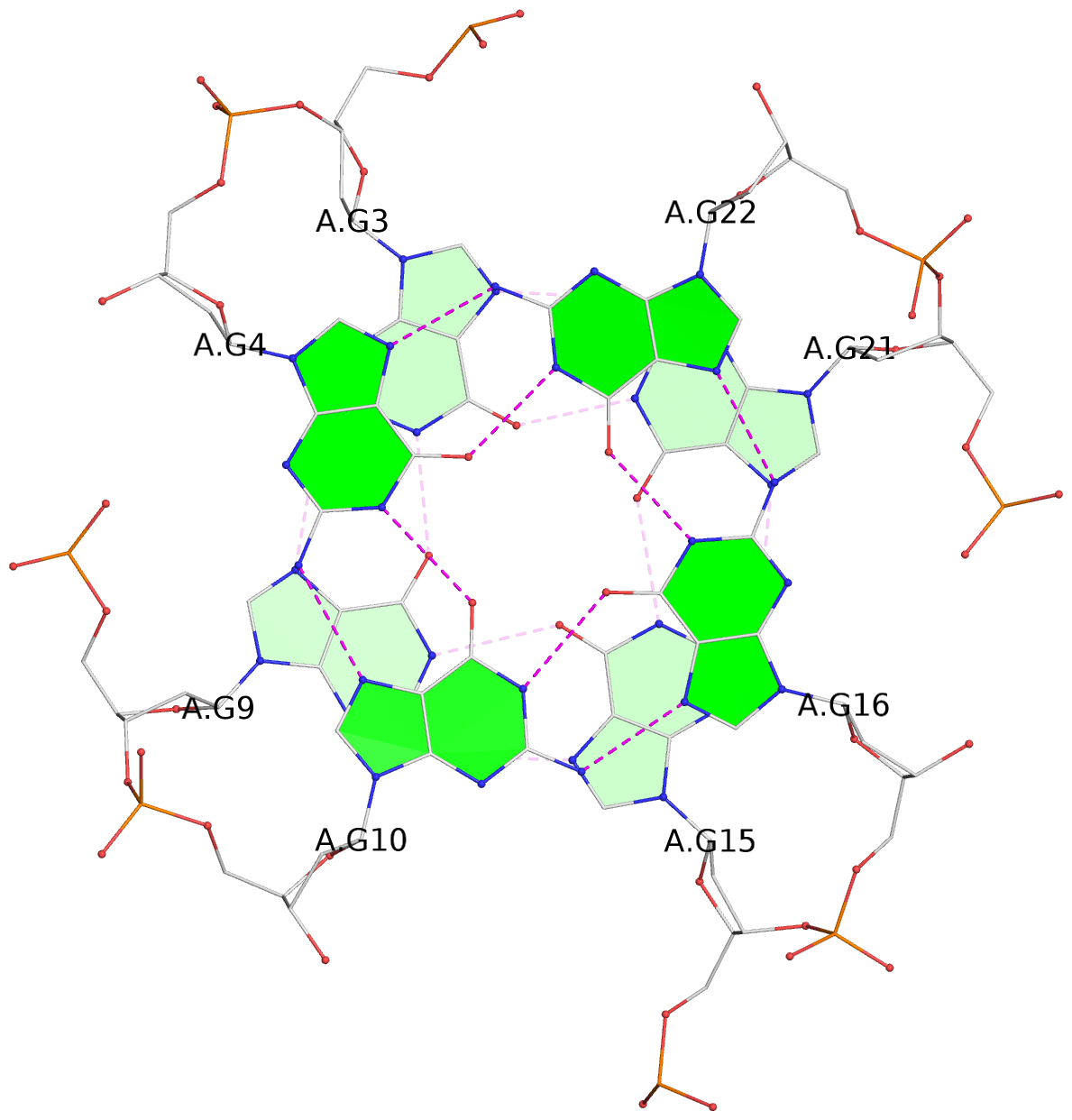
List of 3 G-tetrads
1 glyco-bond=---- sugar=.--- groove=---- planarity=0.225 type=bowl-2 nts=4 GGGG A.DG3,A.DG9,A.DG15,A.DG21 2 glyco-bond=---- sugar=---- groove=---- planarity=0.223 type=bowl-2 nts=4 GGGG A.DG4,A.DG10,A.DG16,A.DG22 3 glyco-bond=---- sugar=---- groove=---- planarity=0.313 type=bowl nts=4 GGGG A.DG5,A.DG11,A.DG17,A.DG23
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.