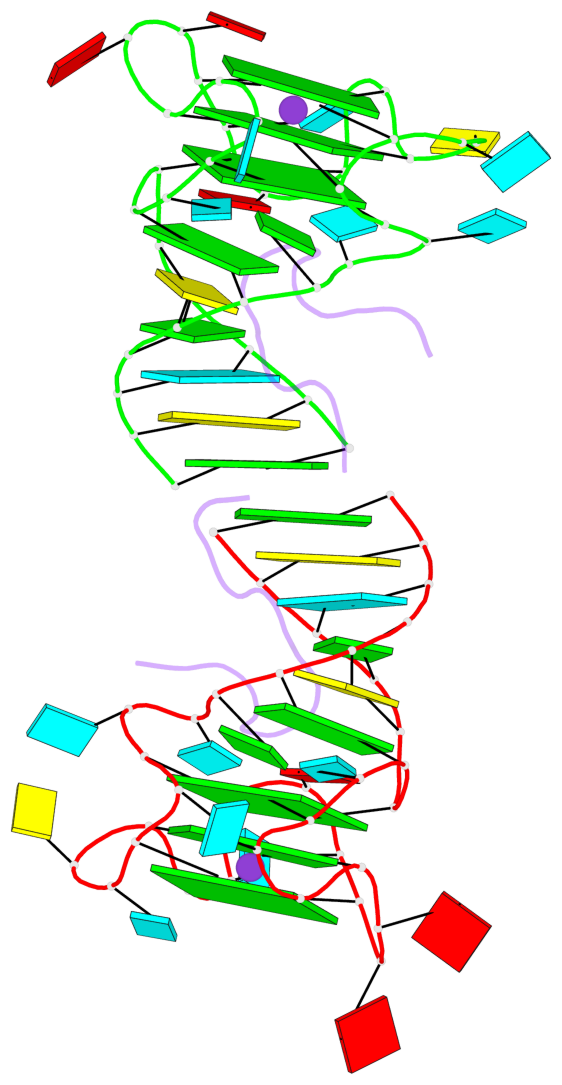
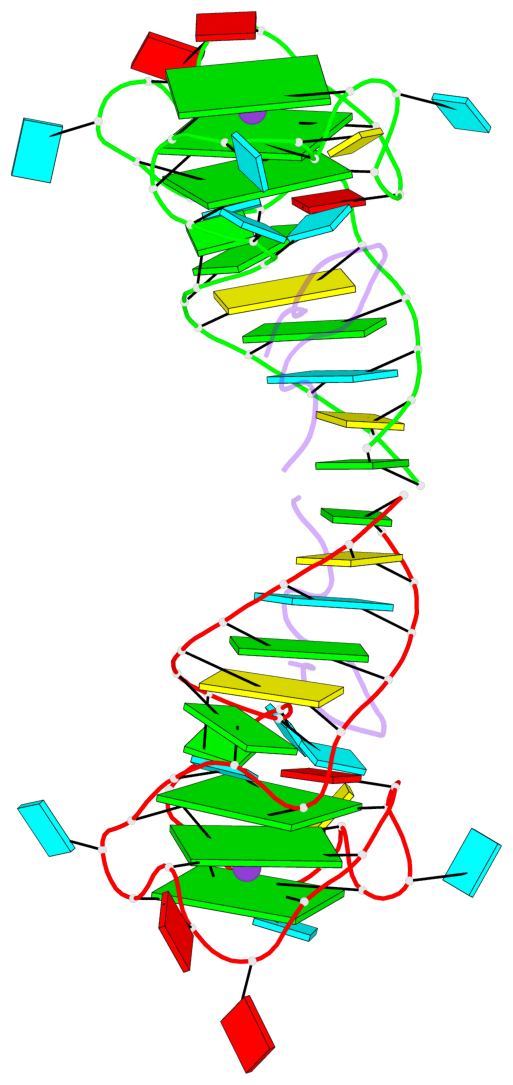
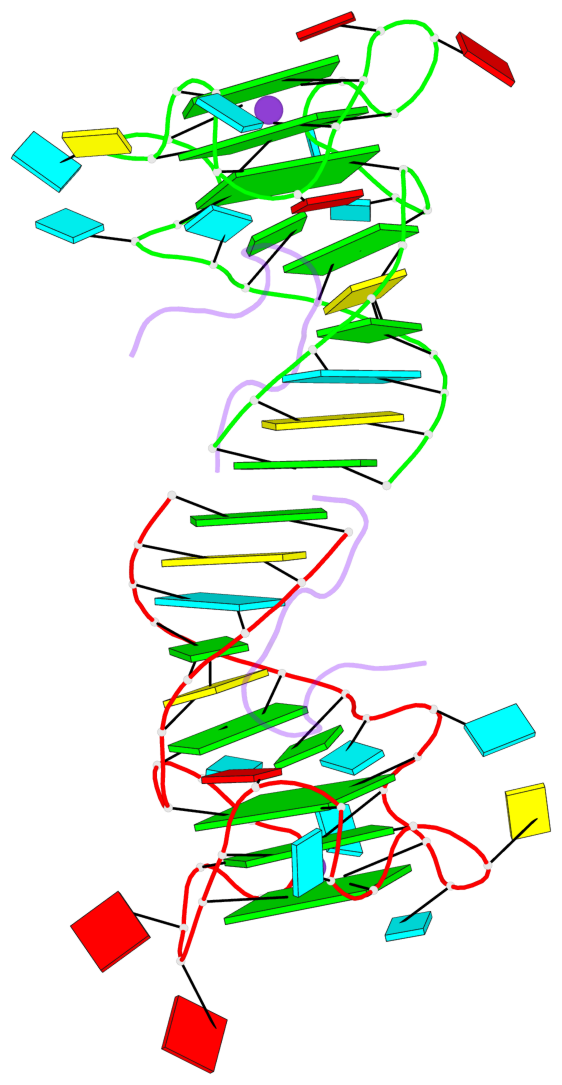
Detailed DSSR results for the G-quadruplex: PDB entry 5de8
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 5de8
- Class
- RNA binding protein-RNA
- Method
- X-ray (3.1 Å)
- Summary
- Crystal structure of the complex between human fmrp rgg motif and g-quadruplex RNA, iridium hexammine bound form.
- Reference
- Vasilyev N, Polonskaia A, Darnell JC, Darnell RB, Patel DJ, Serganov A (2015): "Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP." Proc.Natl.Acad.Sci.USA, 112, E5391-E5400. doi: 10.1073/pnas.1515737112.
- Abstract
- Fragile X Mental Retardation Protein (FMRP) is a regulatory RNA binding protein that plays a central role in the development of several human disorders including Fragile X Syndrome (FXS) and autism. FMRP uses an arginine-glycine-rich (RGG) motif for specific interactions with guanine (G)-quadruplexes, mRNA elements implicated in the disease-associated regulation of specific mRNAs. Here we report the 2.8-Å crystal structure of the complex between the human FMRP RGG peptide bound to the in vitro selected G-rich RNA. In this model system, the RNA adopts an intramolecular K(+)-stabilized G-quadruplex structure composed of three G-quartets and a mixed tetrad connected to an RNA duplex. The RGG peptide specifically binds to the duplex-quadruplex junction, the mixed tetrad, and the duplex region of the RNA through shape complementarity, cation-π interactions, and multiple hydrogen bonds. Many of these interactions critically depend on a type I β-turn, a secondary structure element whose formation was not previously recognized in the RGG motif of FMRP. RNA mutagenesis and footprinting experiments indicate that interactions of the peptide with the duplex-quadruplex junction and the duplex of RNA are equally important for affinity and specificity of the RGG-RNA complex formation. These results suggest that specific binding of cellular RNAs by FMRP may involve hydrogen bonding with RNA duplexes and that RNA duplex recognition can be a characteristic RNA binding feature for RGG motifs in other proteins.
- G4 notes
- 6 G-tetrads, 2 G4 helices, 2 G4 stems, 2(-P-P-P), parallel(4+0), UUUU
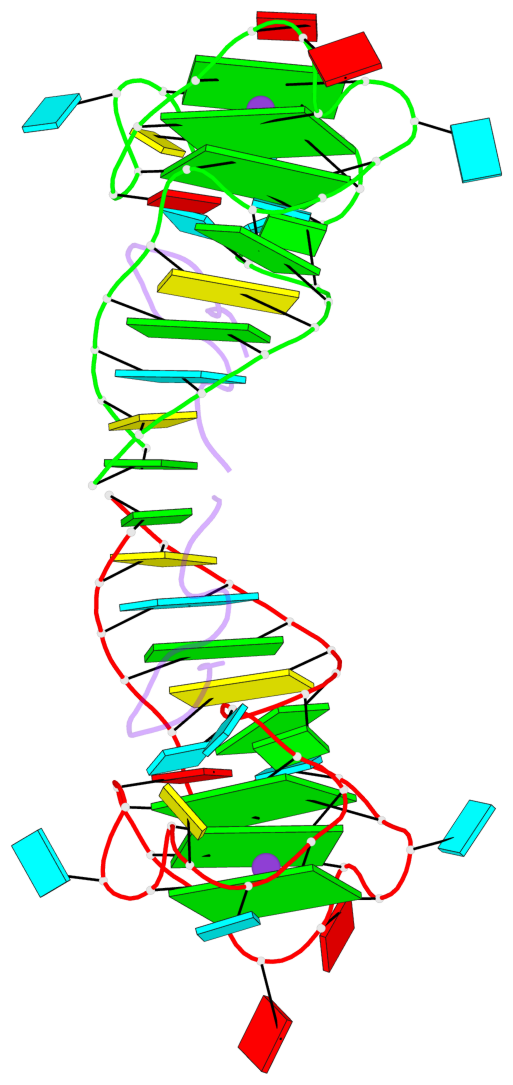
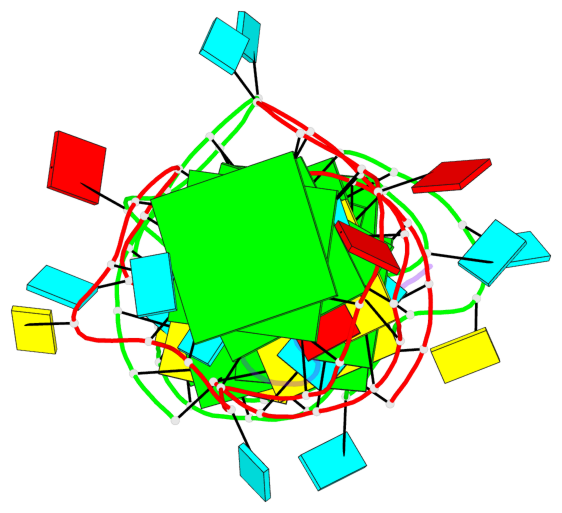
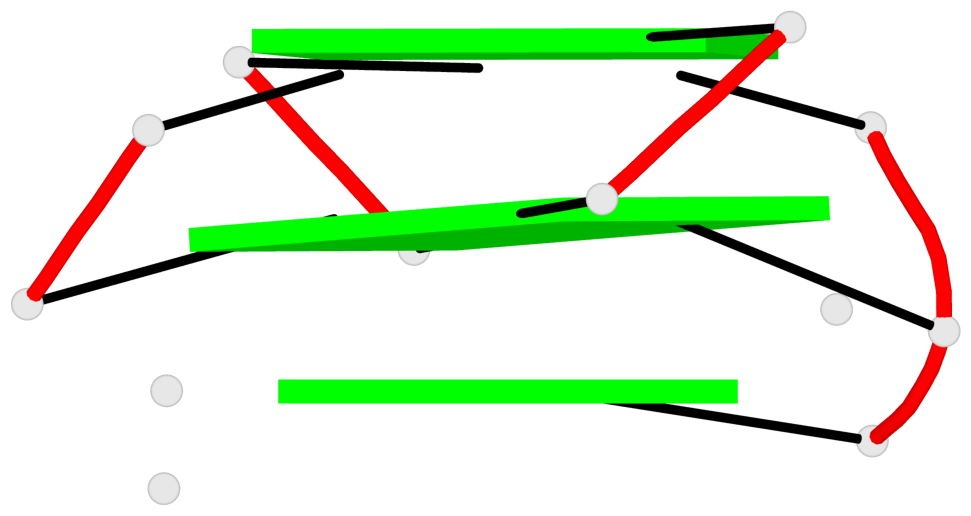
Base-block schematics in six views
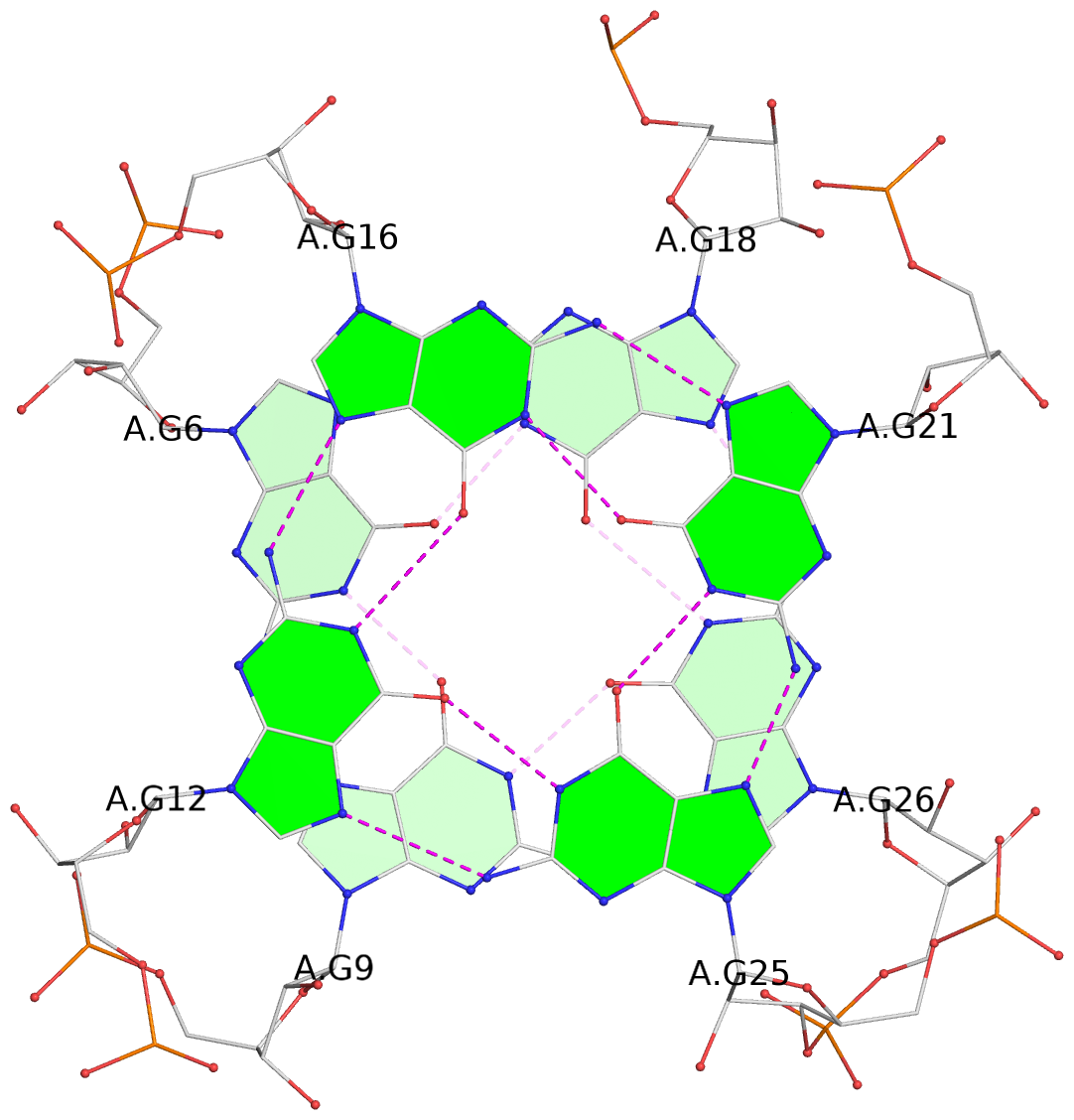
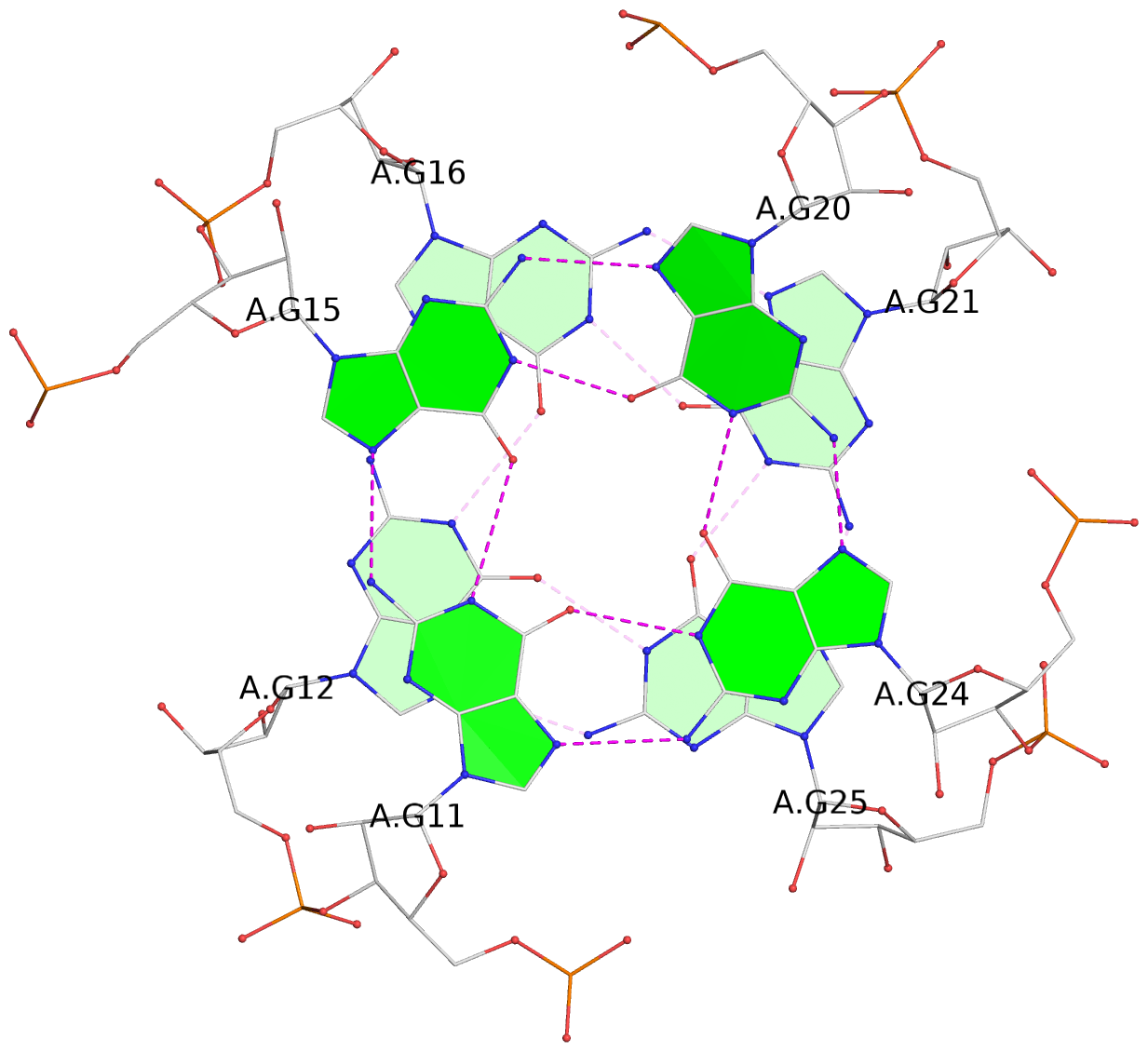
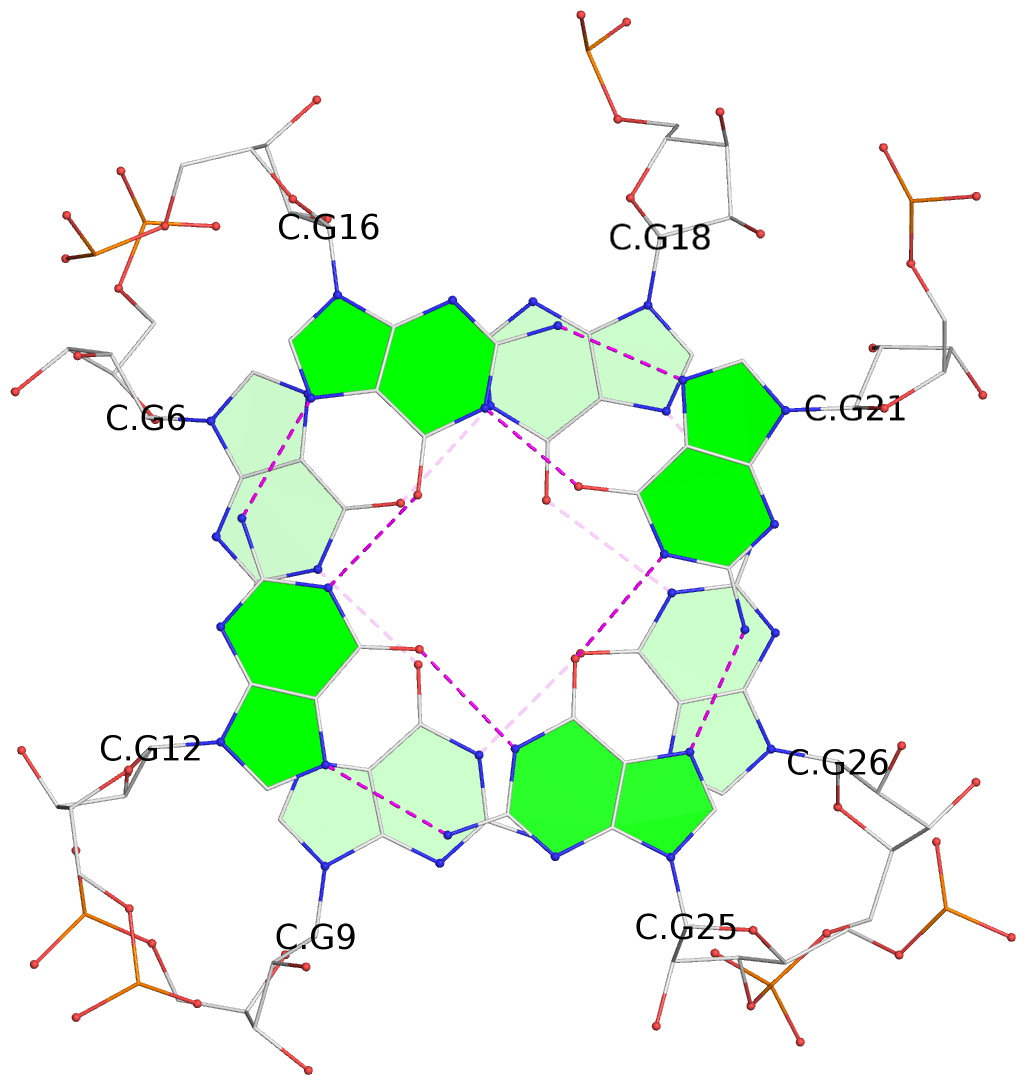
List of 6 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.484 type=other nts=4 GGGG A.G6,A.G9,A.G26,A.G18 2 glyco-bond=---- sugar=3333 groove=---- planarity=0.620 type=other nts=4 GGGG A.G11,A.G15,A.G20,A.G24 3 glyco-bond=---- sugar=---3 groove=---- planarity=0.317 type=saddle nts=4 GGGG A.G12,A.G16,A.G21,A.G25 4 glyco-bond=---- sugar=---- groove=---- planarity=0.484 type=other nts=4 GGGG C.G6,C.G9,C.G26,C.G18 5 glyco-bond=---- sugar=33-3 groove=---- planarity=0.457 type=other nts=4 GGGG C.G11,C.G15,C.G20,C.G24 6 glyco-bond=---- sugar=---3 groove=---- planarity=0.279 type=other nts=4 GGGG C.G12,C.G16,C.G21,C.G25
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
Helix#2, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
Stem#2, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
List of 2 non-stem G4-loops (including the two closing Gs)
1 type=lateral helix=#1 nts=4 GGUG A.G6,A.G7,A.U8,A.G9 2 type=lateral helix=#2 nts=4 GGUG C.G6,C.G7,C.U8,C.G9