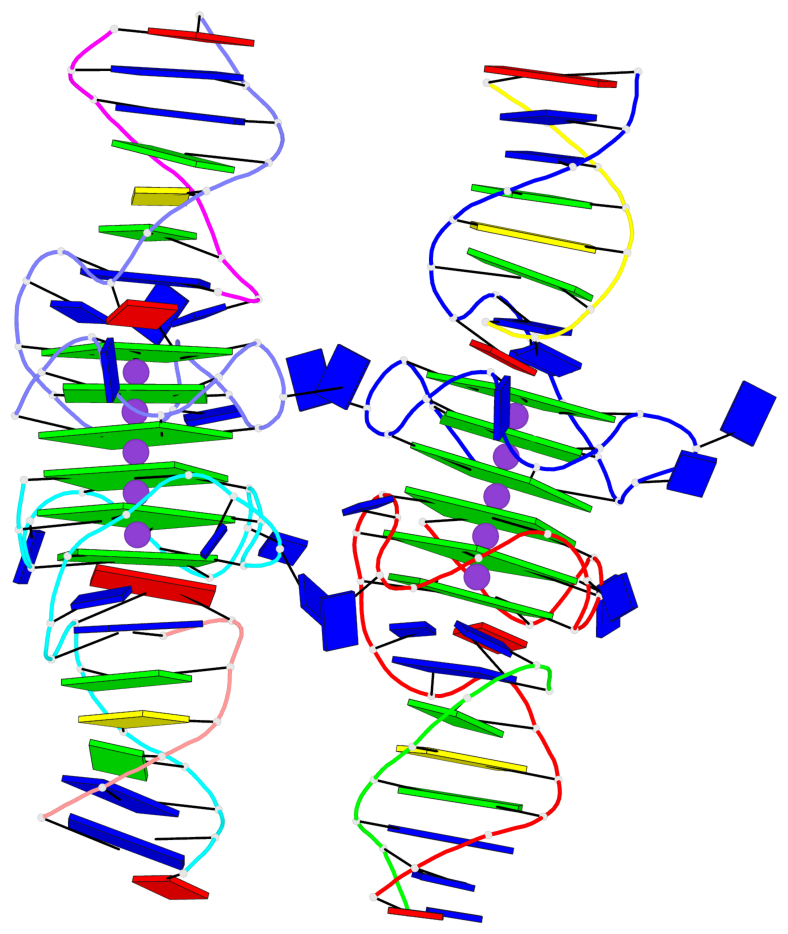
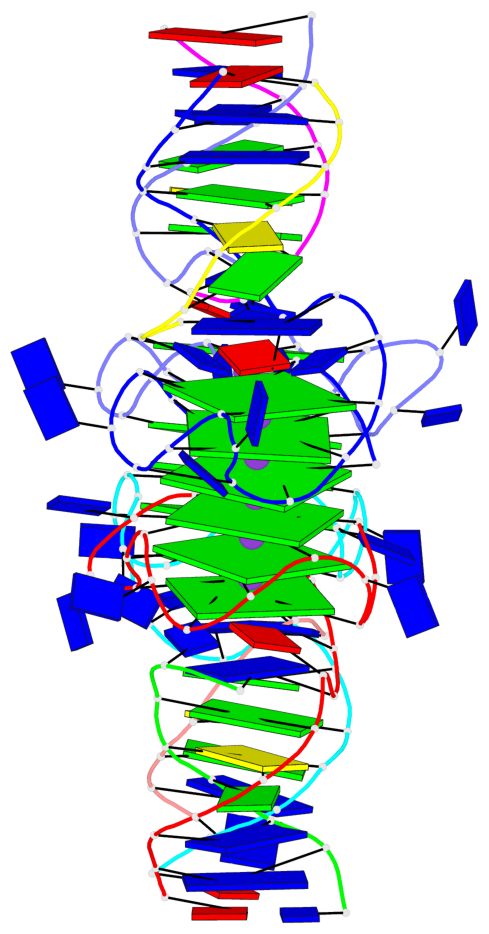
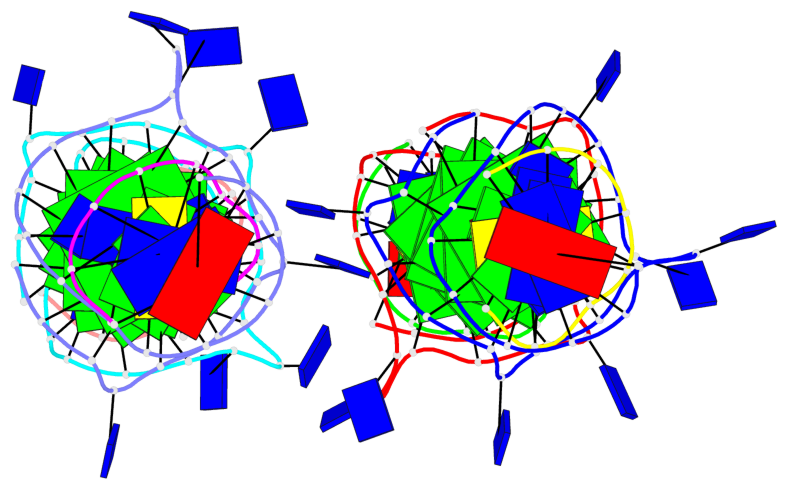
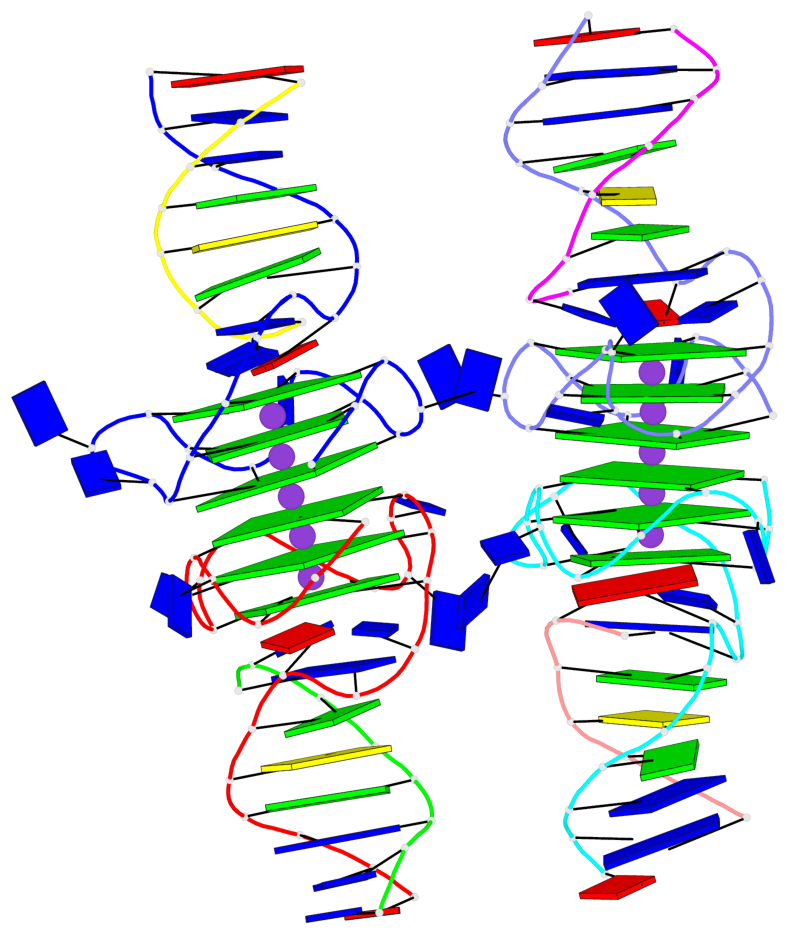
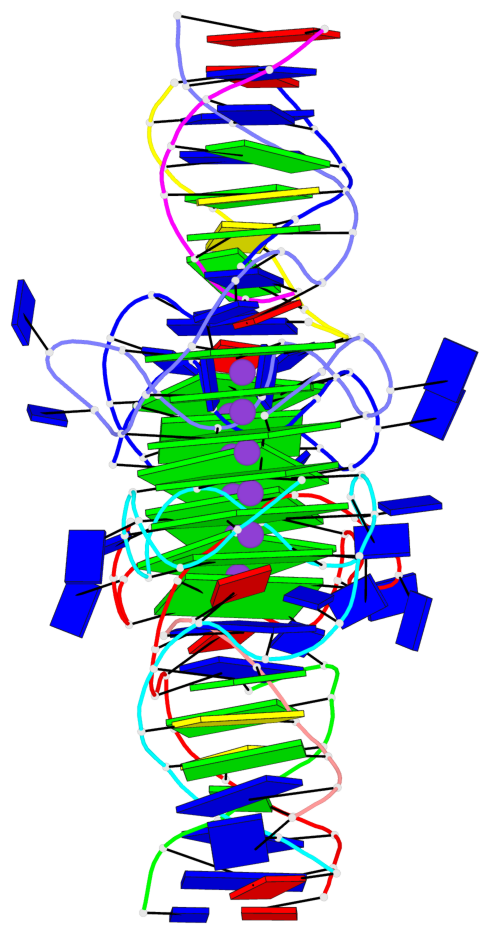
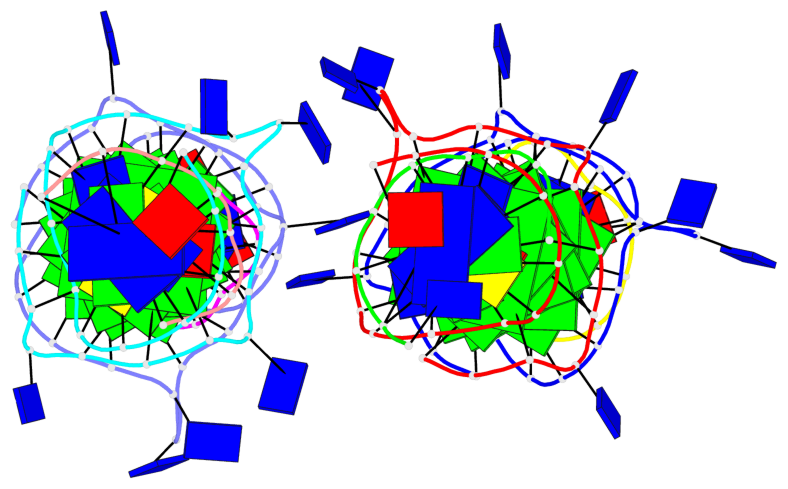
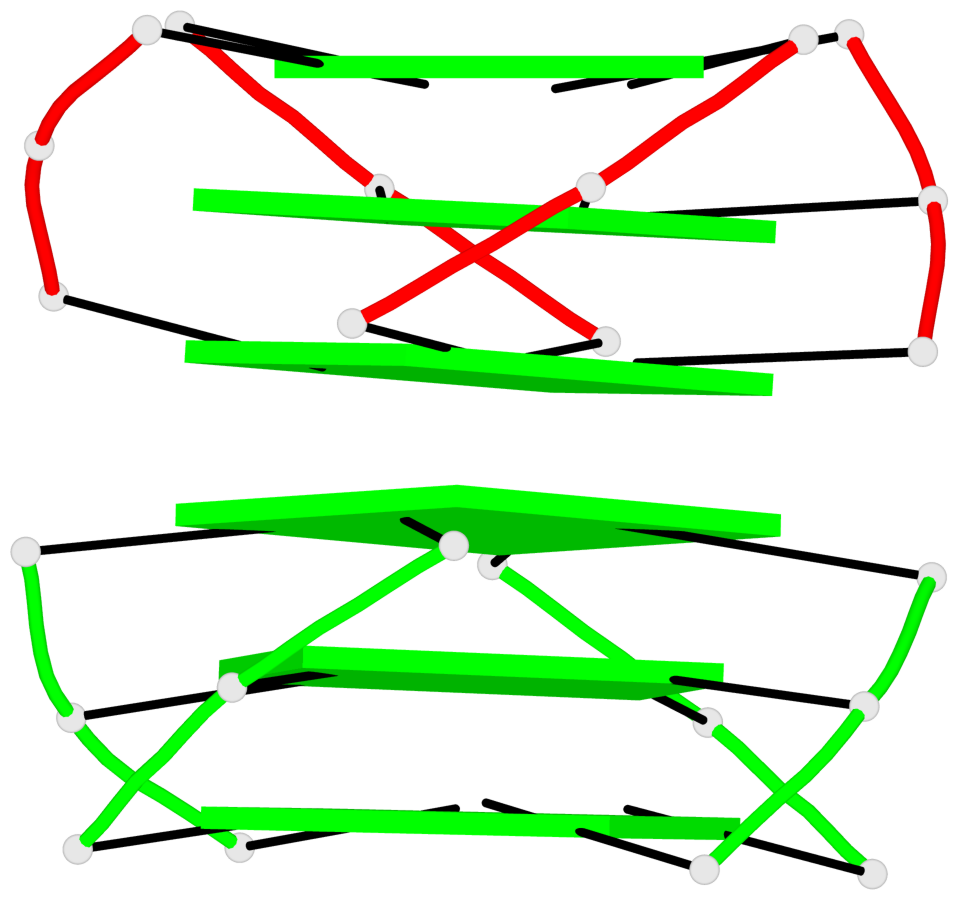
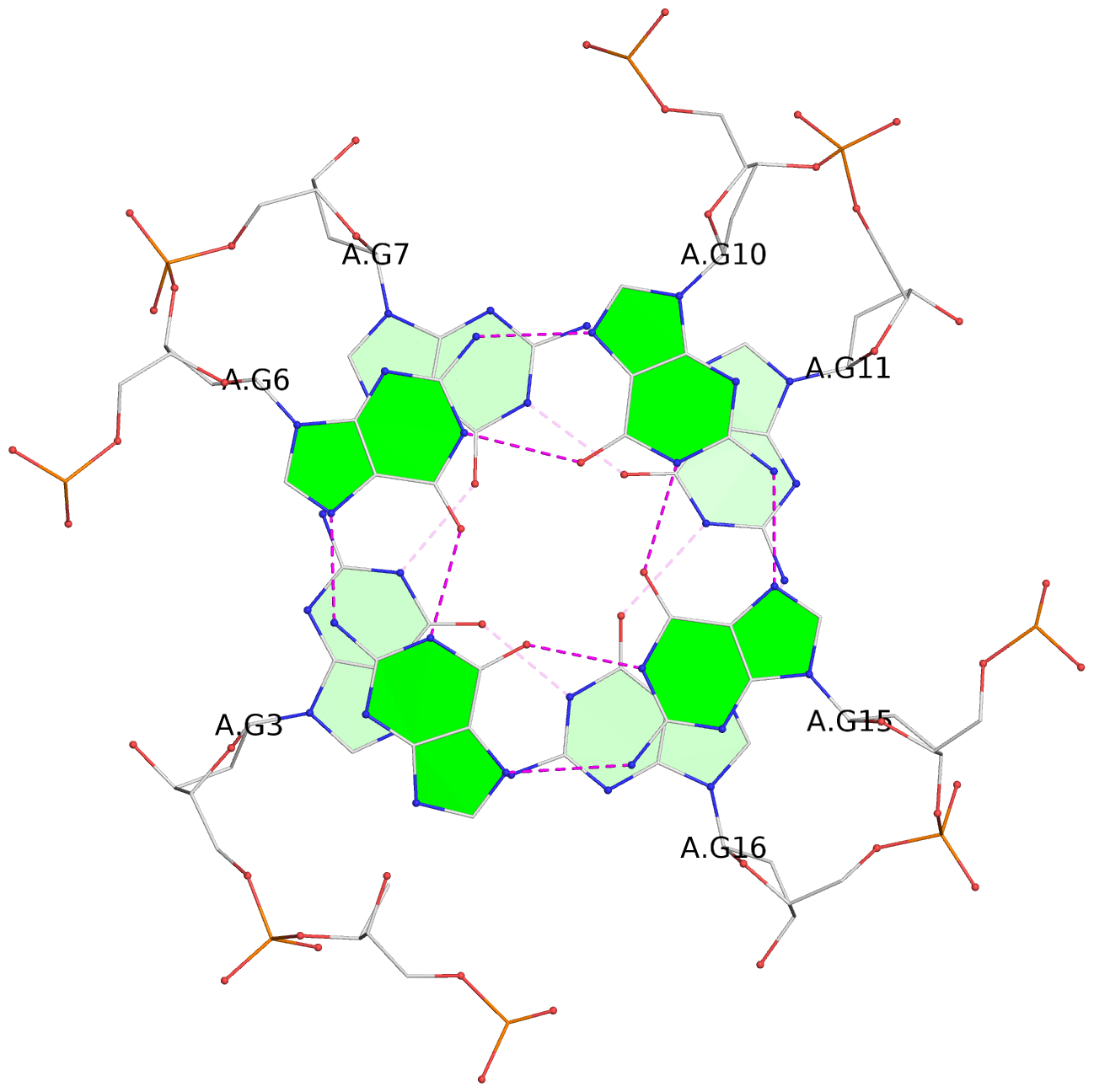
Detailed DSSR results for the G-quadruplex: PDB entry 5dww
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 5dww
- Class
- DNA
- Method
- X-ray (2.79 Å)
- Summary
- Structural insights into the quadruplex-duplex 3' interface formed from a telomeric repeat - ttloop
- Reference
- Russo Krauss I, Ramaswamy S, Neidle S, Haider S, Parkinson GN (2016): "Structural Insights into the Quadruplex-Duplex 3' Interface Formed from a Telomeric Repeat: A Potential Molecular Target." J.Am.Chem.Soc., 138, 1226-1233. doi: 10.1021/jacs.5b10492.
- Abstract
- We report here on an X-ray crystallographic and molecular modeling investigation into the complex 3' interface formed between putative parallel stranded G-quadruplexes and a duplex DNA sequence constructed from the human telomeric repeat sequence TTAGGG. Our crystallographic approach provides a detailed snapshot of a telomeric 3' quadruplex-duplex junction: a junction that appears to have the potential to form a unique molecular target for small molecule binding and interference with telomere-related functions. This unique target is particularly relevant as current high-affinity compounds that bind putative G-quadruplex forming sequences only rarely have a high degree of selectivity for a particular quadruplex. Here DNA junctions were assembled using different putative quadruplex-forming scaffolds linked at the 3' end to a telomeric duplex sequence and annealed to a complementary strand. We successfully generated a series of G-quadruplex-duplex containing crystals, both alone and in the presence of ligands. The structures demonstrate the formation of a parallel folded G-quadruplex and a B-form duplex DNA stacked coaxially. Most strikingly, structural data reveals the consistent formation of a TAT triad platform between the two motifs. This triad allows for a continuous stack of bases to link the quadruplex motif with the duplex region. For these crystal structures formed in the absence of ligands, the TAT triad interface occludes ligand binding at the 3' quadruplex-duplex interface, in agreement with in silico docking predictions. However, with the rearrangement of a single nucleotide, a stable pocket can be produced, thus providing an opportunity for the binding of selective molecules at the interface.
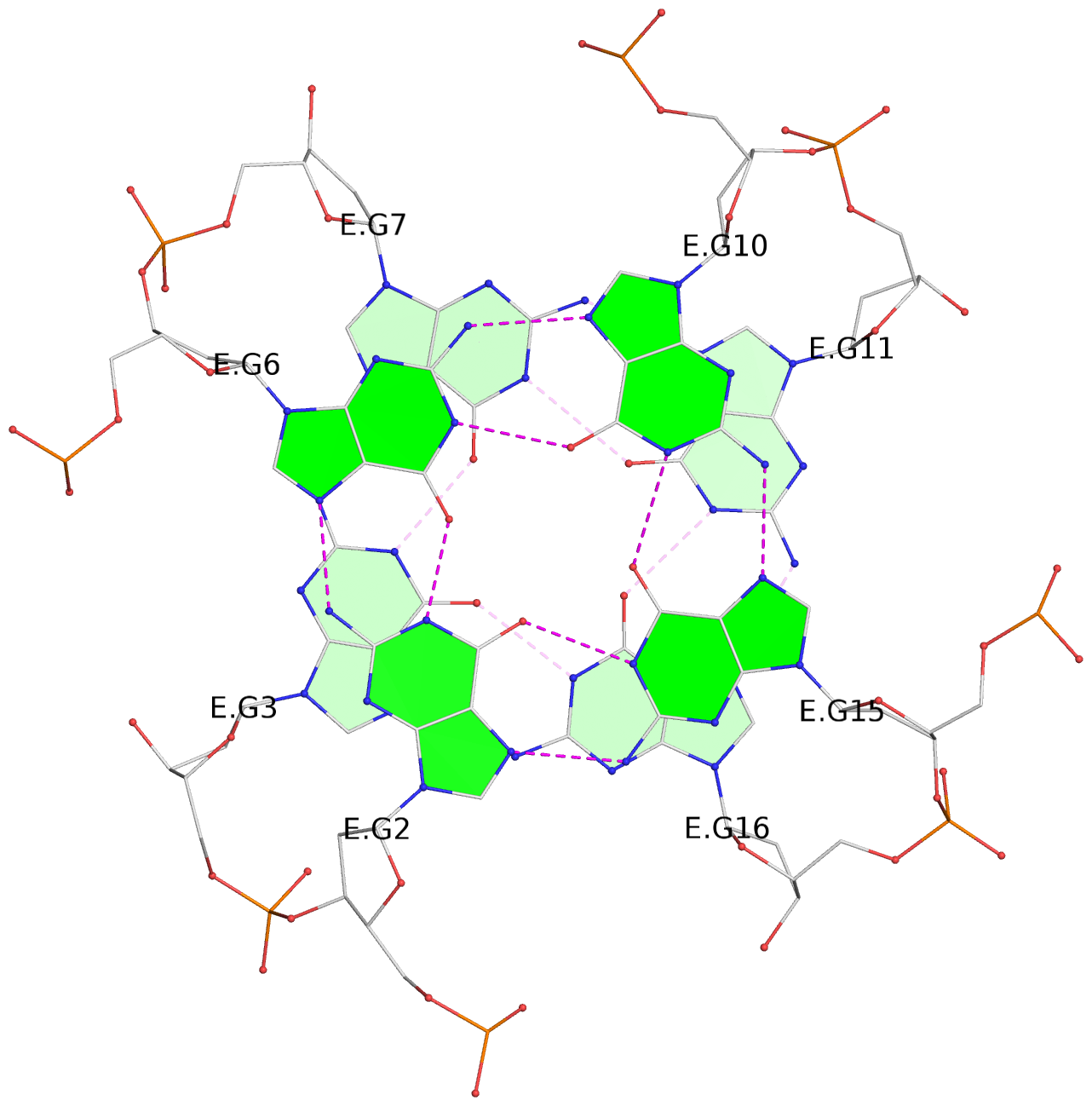
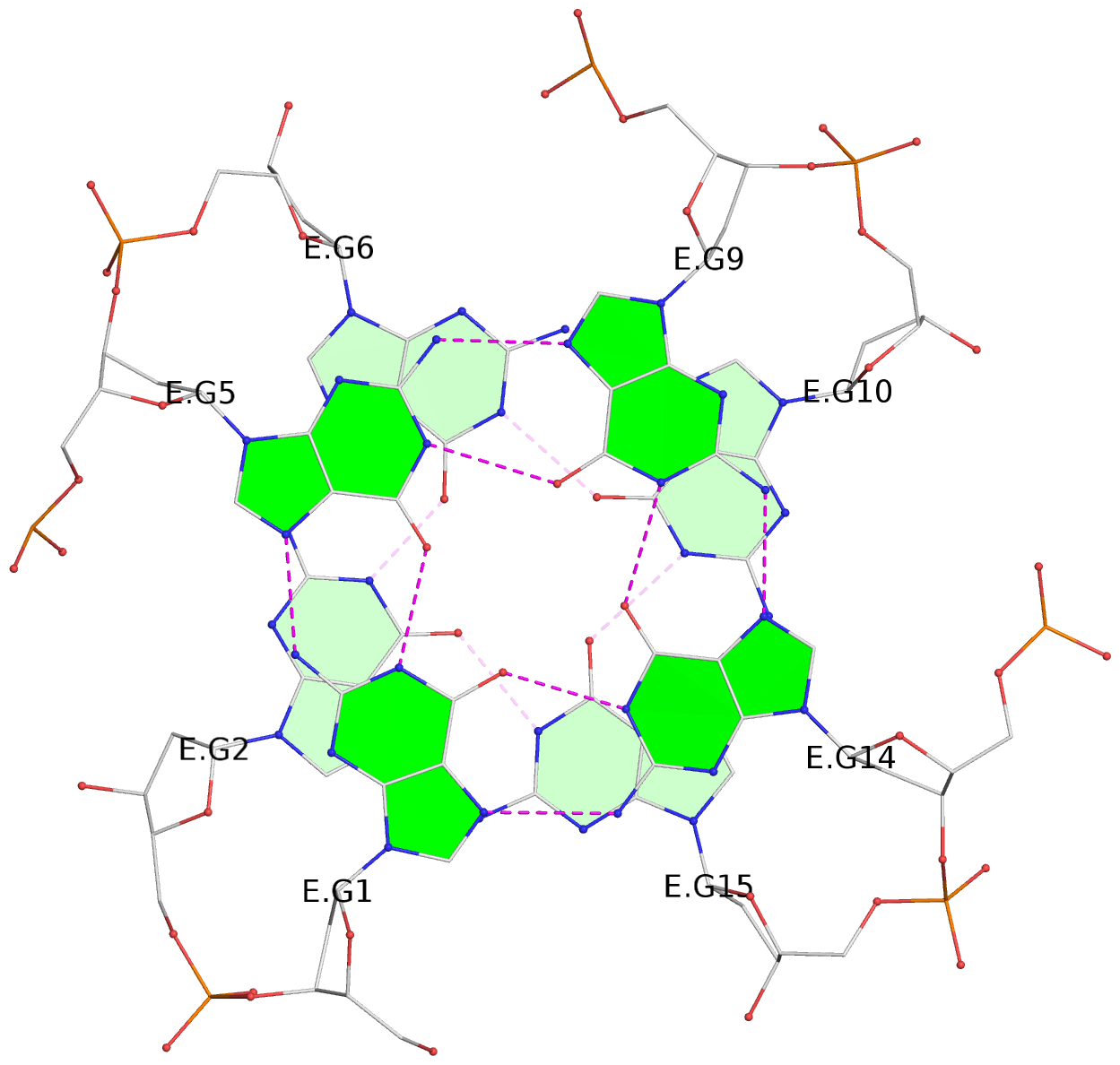
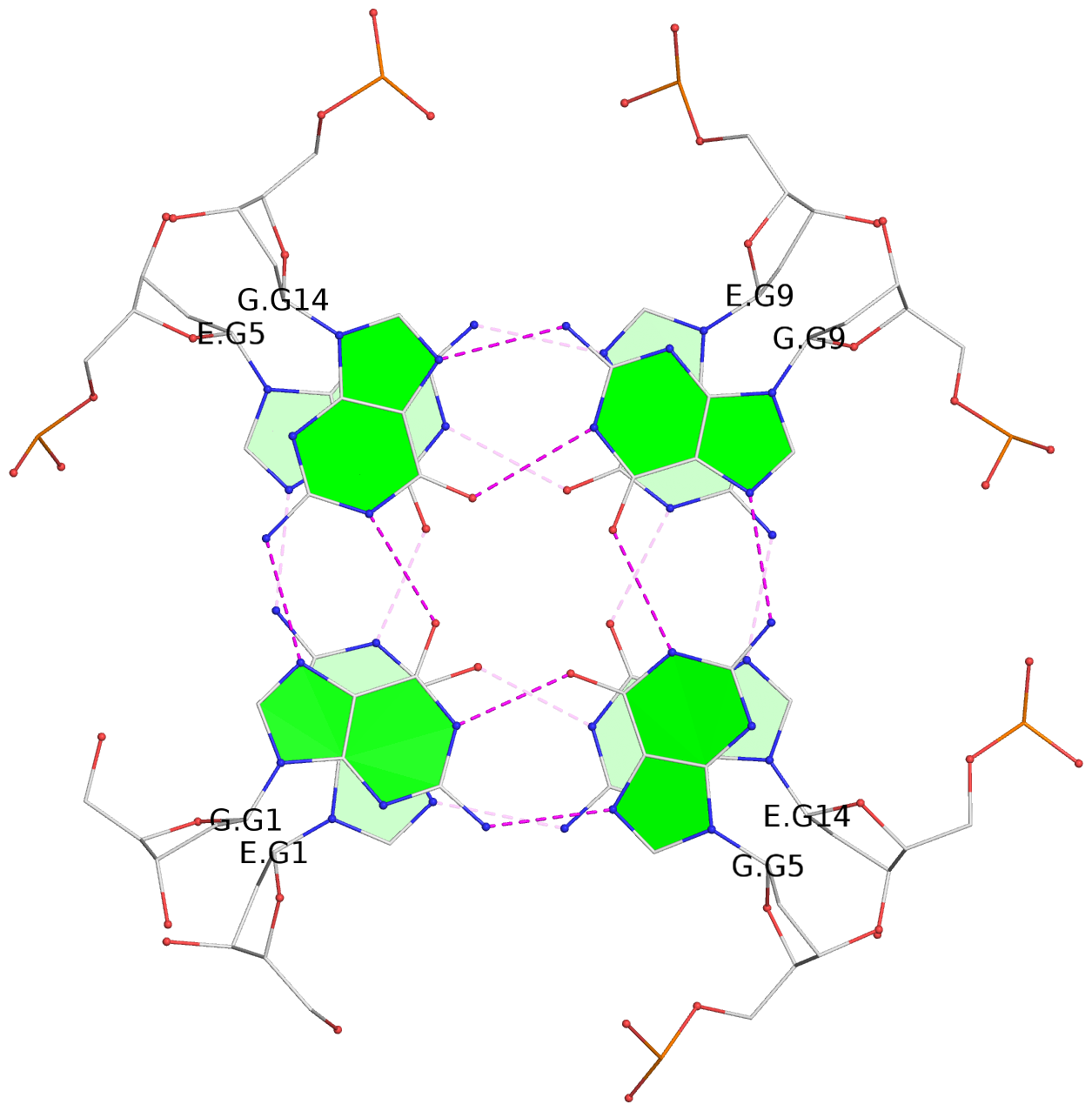
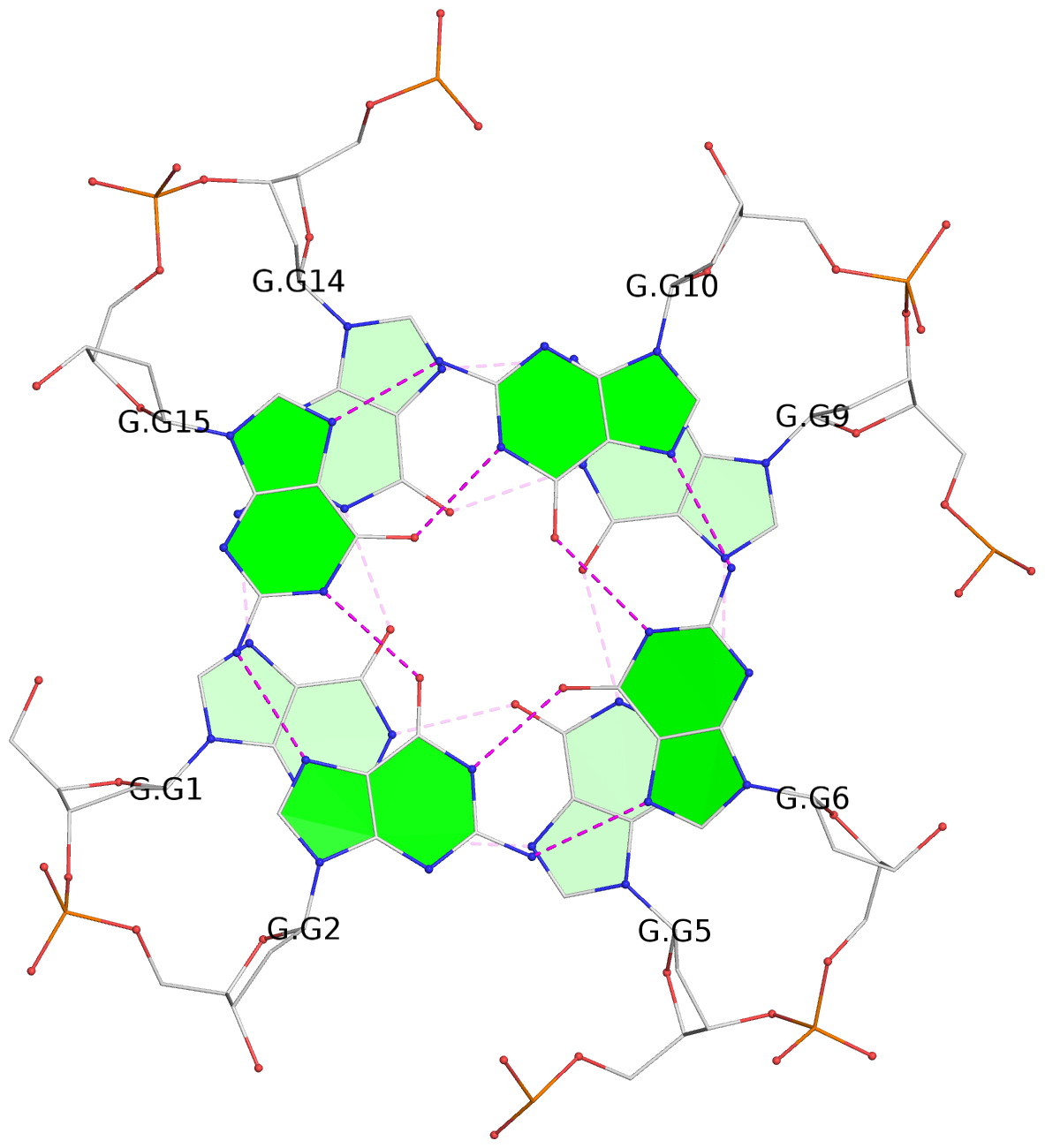
- G4 notes
- 12 G-tetrads, 2 G4 helices, 4 G4 stems, 2 G4 coaxial stacks, 3(-P-P-P), parallel(4+0), UUUU, coaxial interfaces: 5'/5'
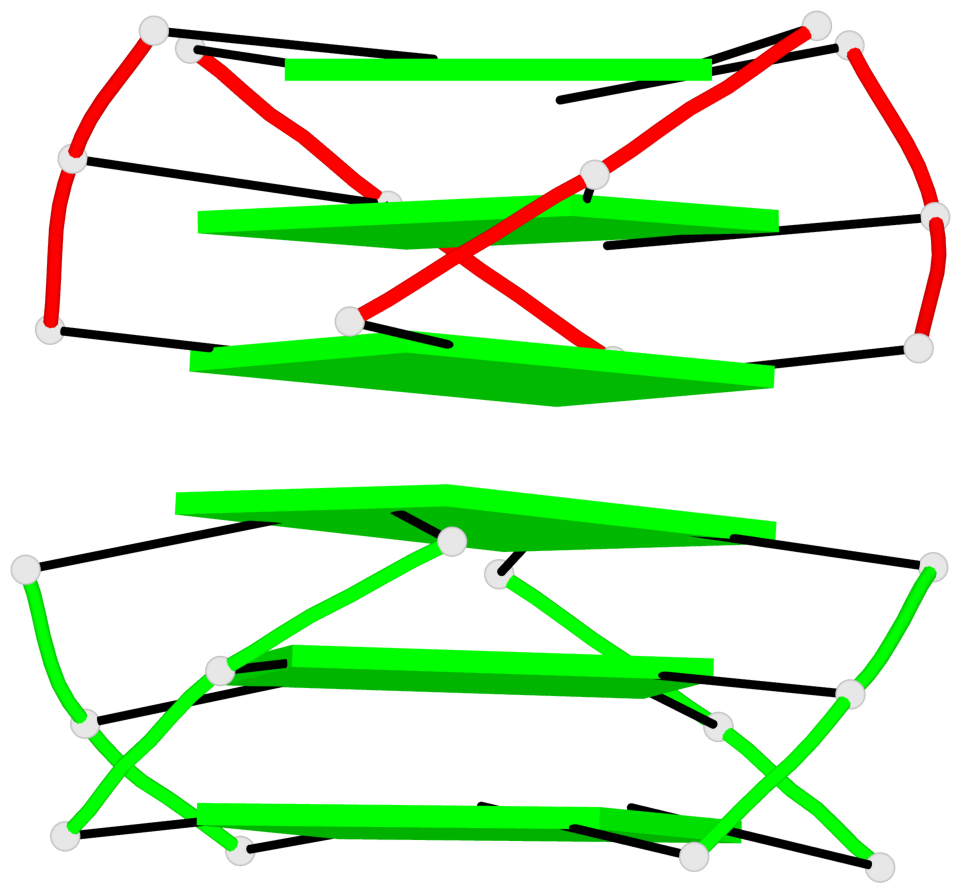
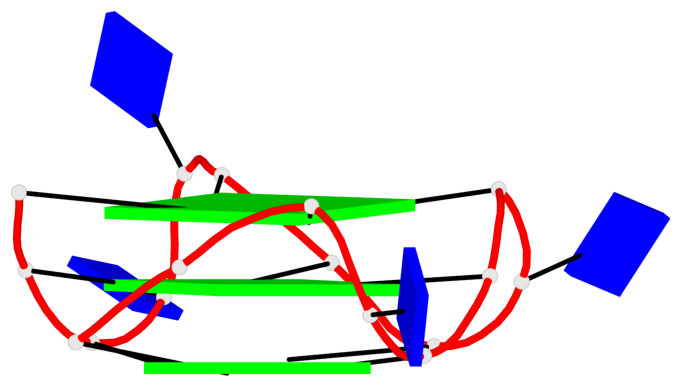
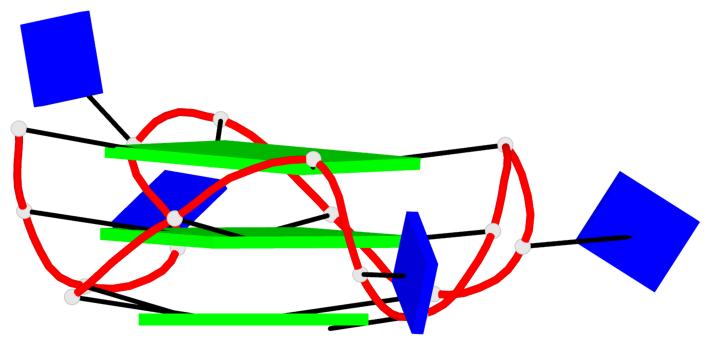
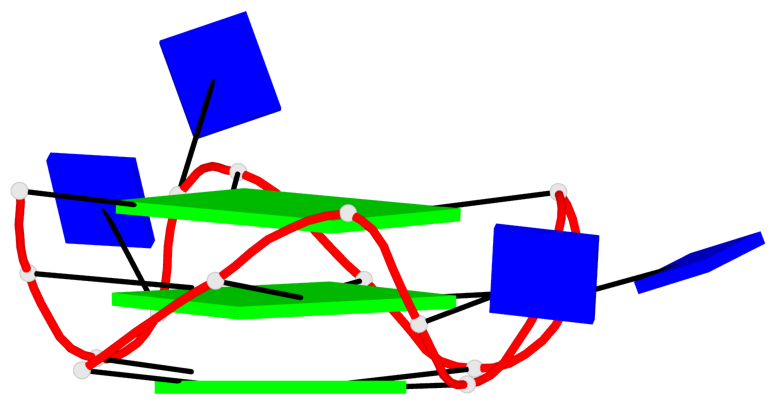
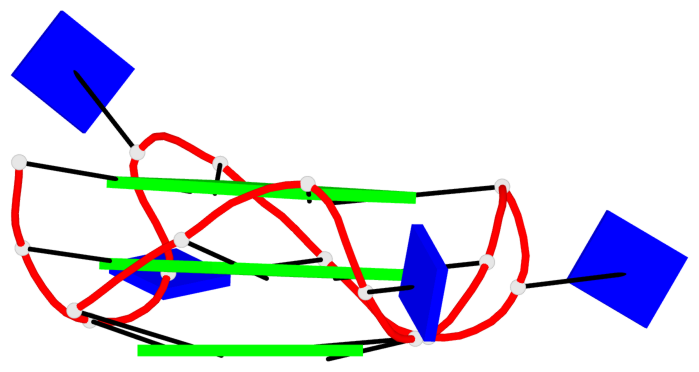
Base-block schematics in six views
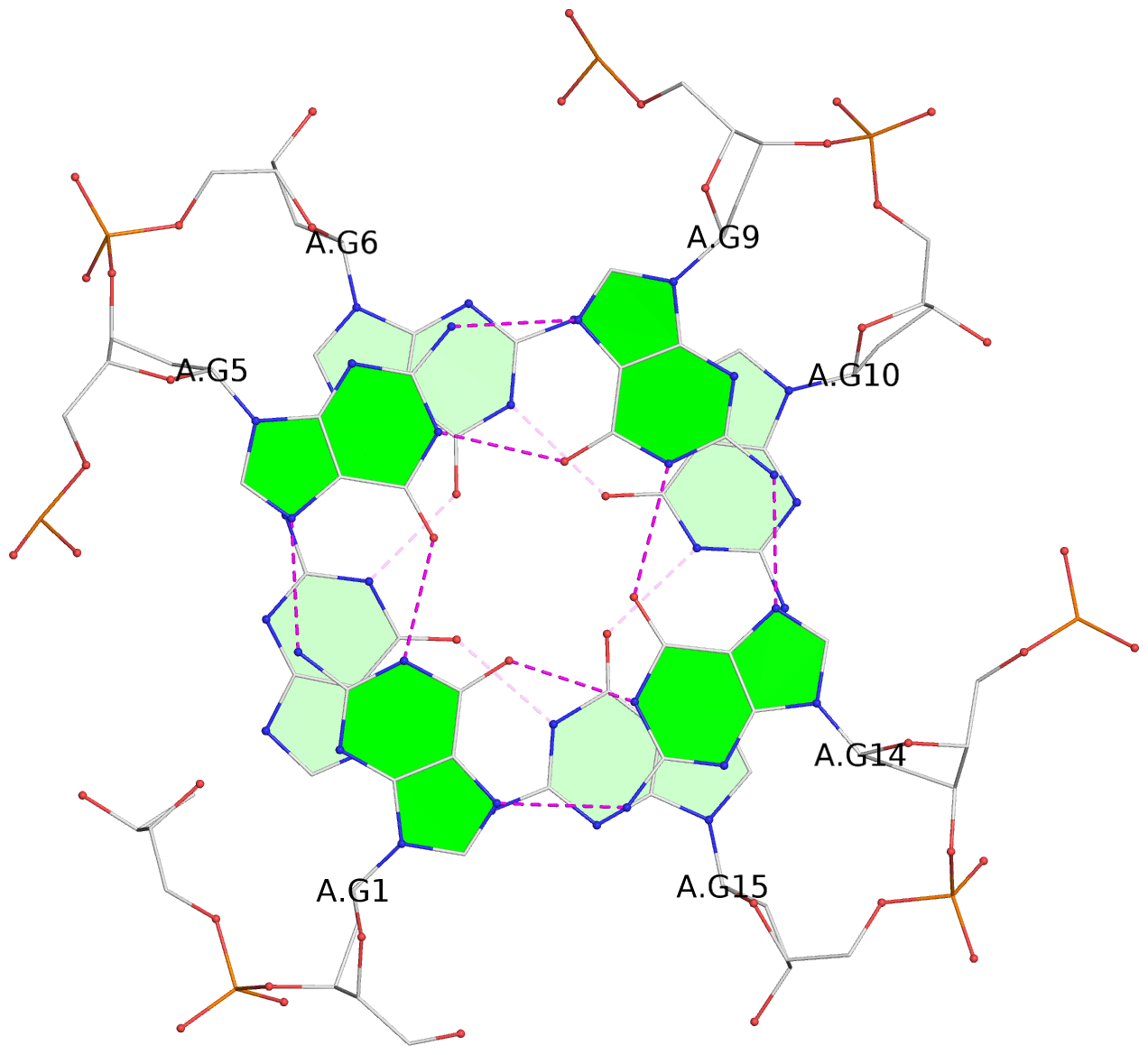
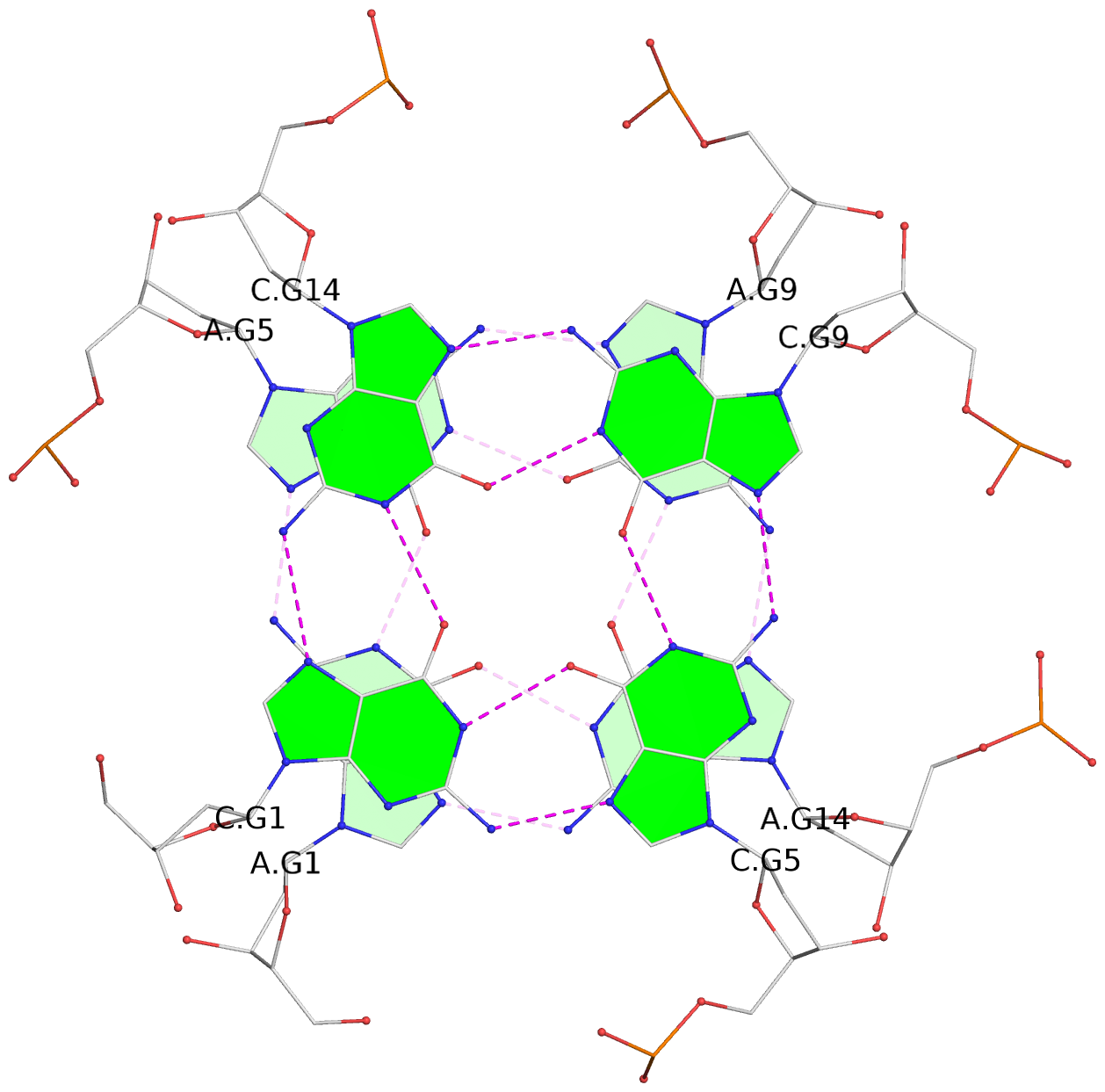
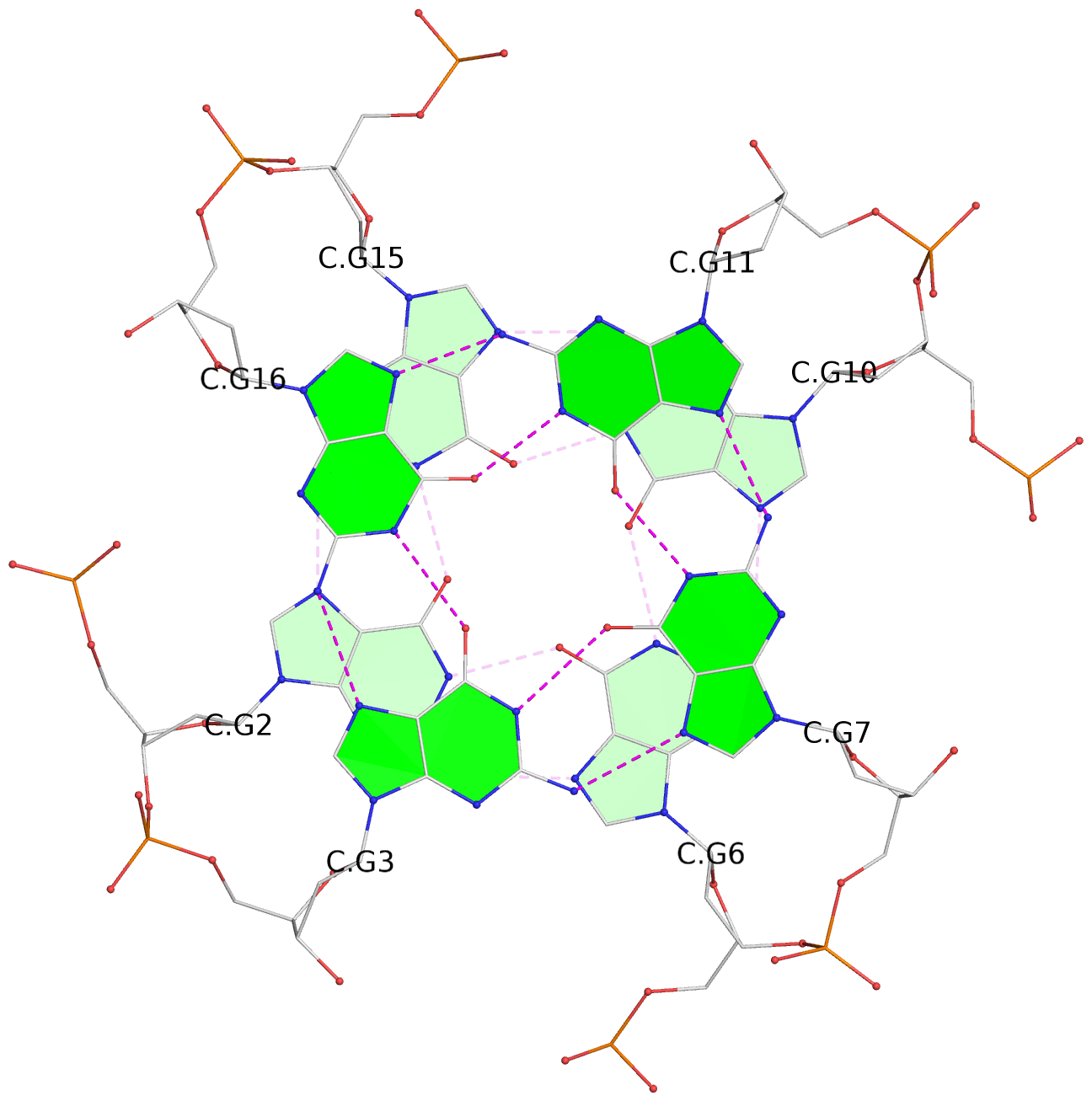
List of 12 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.138 type=planar nts=4 GGGG A.DG1,A.DG5,A.DG9,A.DG14 2 glyco-bond=---- sugar=.--- groove=---- planarity=0.120 type=planar nts=4 GGGG A.DG2,A.DG6,A.DG10,A.DG15 3 glyco-bond=---- sugar=---- groove=---- planarity=0.268 type=bowl nts=4 GGGG A.DG3,A.DG7,A.DG11,A.DG16 4 glyco-bond=---- sugar=--.. groove=---- planarity=0.130 type=planar nts=4 GGGG C.DG1,C.DG5,C.DG9,C.DG14 5 glyco-bond=---- sugar=---- groove=---- planarity=0.156 type=planar nts=4 GGGG C.DG2,C.DG6,C.DG10,C.DG15 6 glyco-bond=---- sugar=---- groove=---- planarity=0.306 type=bowl nts=4 GGGG C.DG3,C.DG7,C.DG11,C.DG16 7 glyco-bond=---- sugar=---- groove=---- planarity=0.179 type=other nts=4 GGGG E.DG1,E.DG5,E.DG9,E.DG14 8 glyco-bond=---- sugar=..-- groove=---- planarity=0.246 type=other nts=4 GGGG E.DG2,E.DG6,E.DG10,E.DG15 9 glyco-bond=---- sugar=-.-- groove=---- planarity=0.227 type=other nts=4 GGGG E.DG3,E.DG7,E.DG11,E.DG16 10 glyco-bond=---- sugar=---- groove=---- planarity=0.204 type=other nts=4 GGGG G.DG1,G.DG5,G.DG9,G.DG14 11 glyco-bond=---- sugar=---- groove=---- planarity=0.249 type=other nts=4 GGGG G.DG2,G.DG6,G.DG10,G.DG15 12 glyco-bond=---- sugar=.--- groove=---- planarity=0.292 type=bowl nts=4 GGGG G.DG3,G.DG7,G.DG11,G.DG16
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 6 G-tetrad layers, inter-molecular, with 2 stems
Helix#2, 6 G-tetrad layers, inter-molecular, with 2 stems
List of 4 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 3(-P-P-P), parallel(4+0)
Stem#2, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 3(-P-P-P), parallel(4+0)
Stem#3, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 3(-P-P-P), parallel(4+0)
Stem#4, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 3(-P-P-P), parallel(4+0)
List of 2 G4 coaxial stacks
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [5'/5'] 2 G4 helix#2 contains 2 G4 stems: [#3,#4] [5'/5']