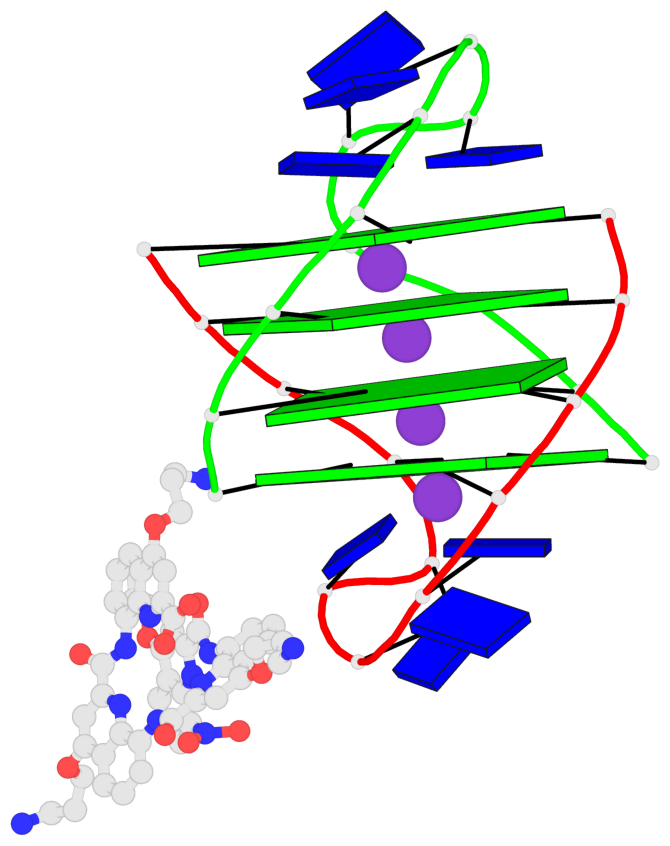
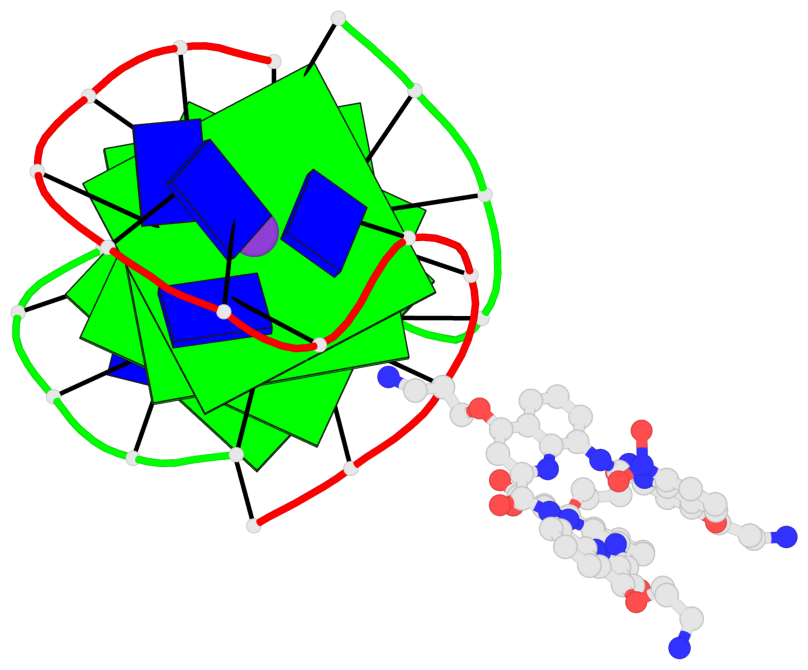
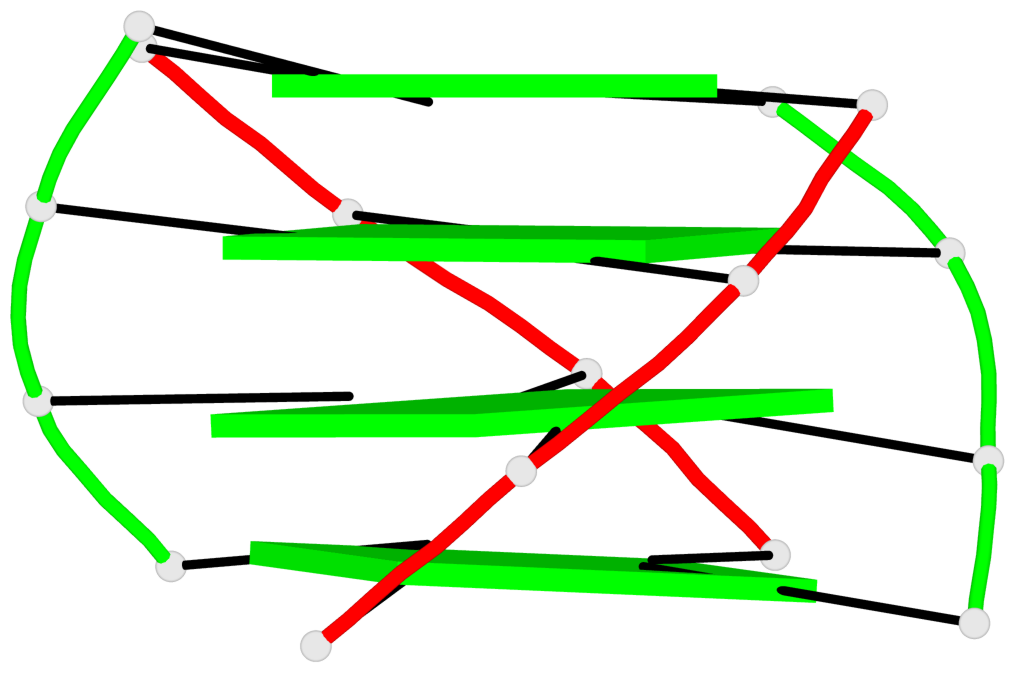
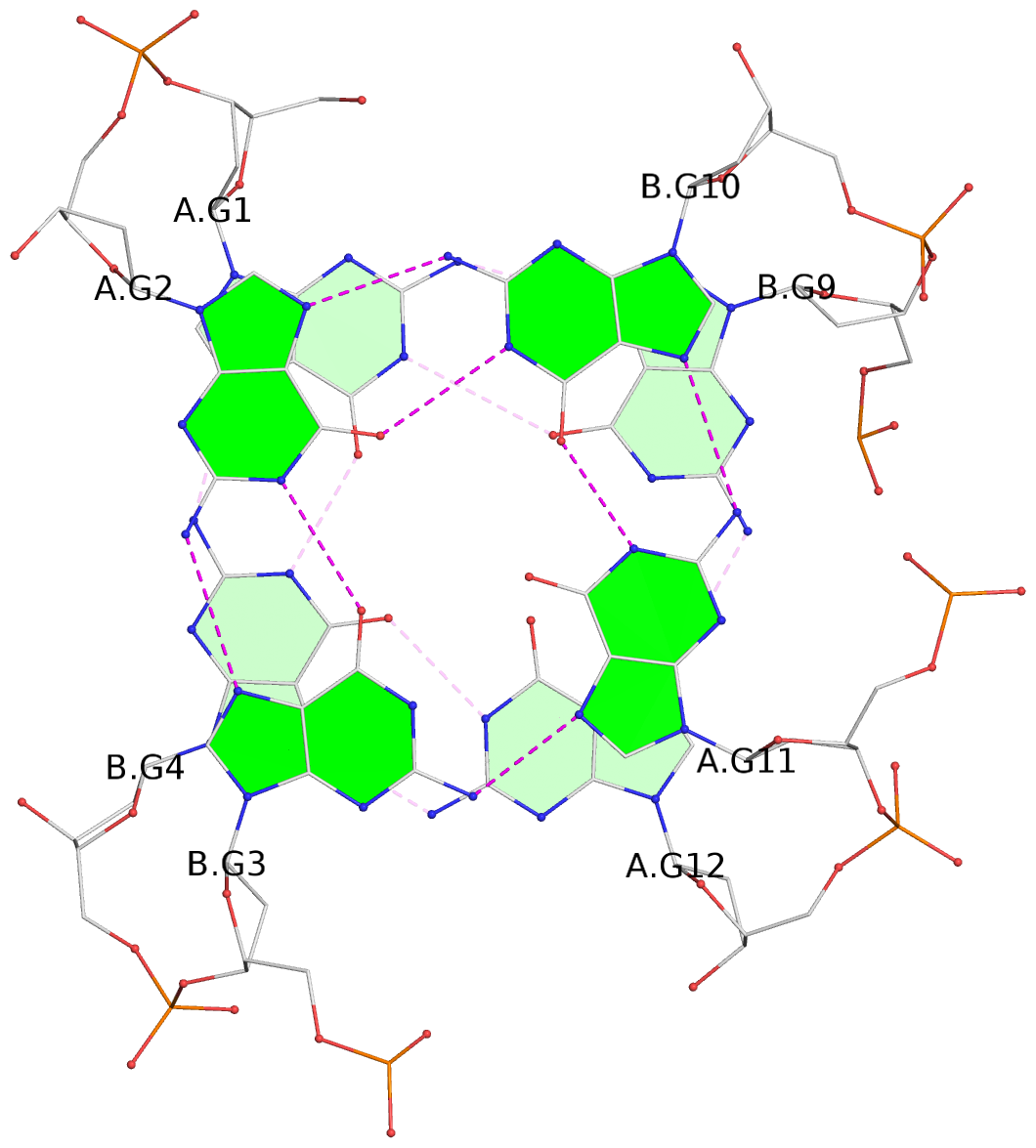
Detailed DSSR results for the G-quadruplex: PDB entry 5hix
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 5hix
- Class
- DNA
- Method
- X-ray (2.48 Å)
- Summary
- Cocrystal structure of an anti-parallel DNA g-quadruplex and a tetra-quinoline foldamer
- Reference
- Mandal PK, Baptiste B, Langlois d'Estaintot B, Kauffmann B, Huc I (2016): "Multivalent Interactions between an Aromatic Helical Foldamer and a DNA G-Quadruplex in the Solid State." Chembiochem, 17, 1911-1914. doi: 10.1002/cbic.201600281.
- Abstract
- Quinoline-based oligoamide foldamers have been identified as a potent class of ligands for G-quadruplex DNA. Their helical structure is thought to target G-quadruplex loops or grooves and not G-tetrads. We report a co-crystal structure of the antiparallel hairpin dimeric DNA G-quadruplex (G4 T4 G4 )2 with tetramer 1-a helically folded oligo-quinolinecarboxamide bearing cationic side chains-that is consistent with this hypothesis. Multivalent foldamer-DNA interactions that modify the packing of (G4 T4 G4 )2 in the solid state are observed.
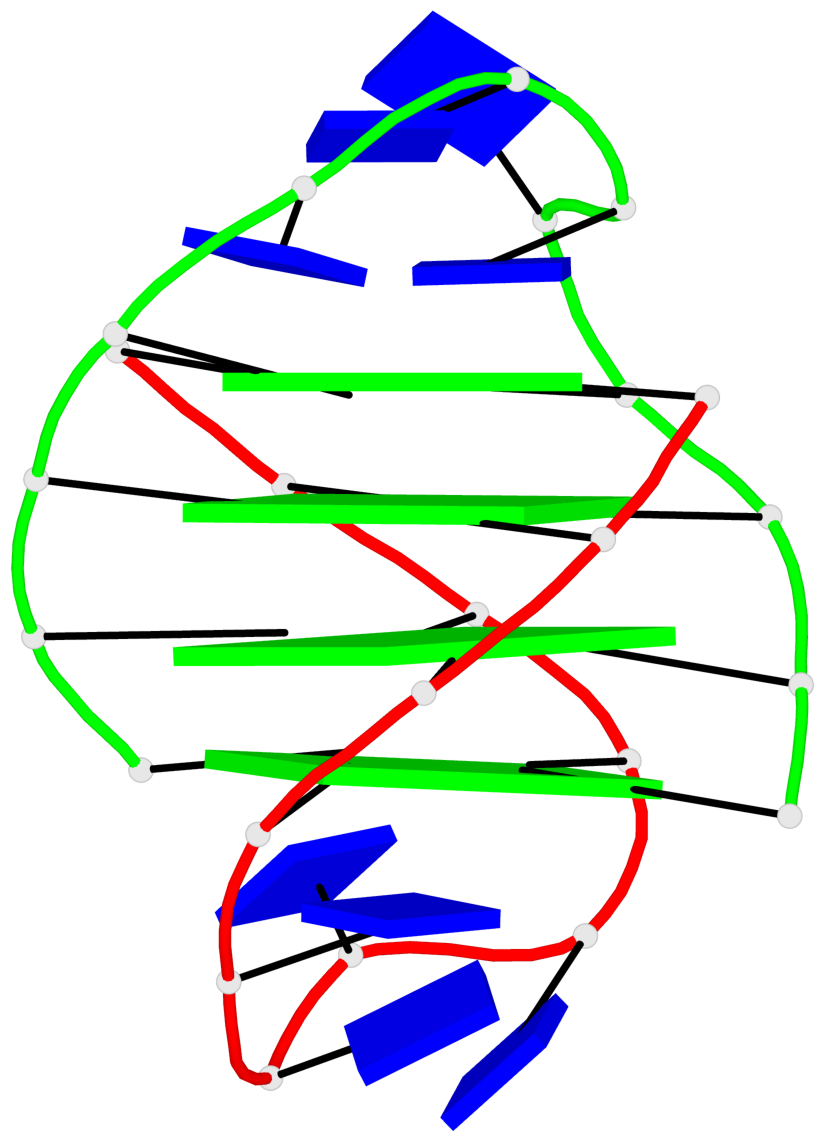
- G4 notes
- 4 G-tetrads, 1 G4 helix, 1 G4 stem, (2+2), UDDU
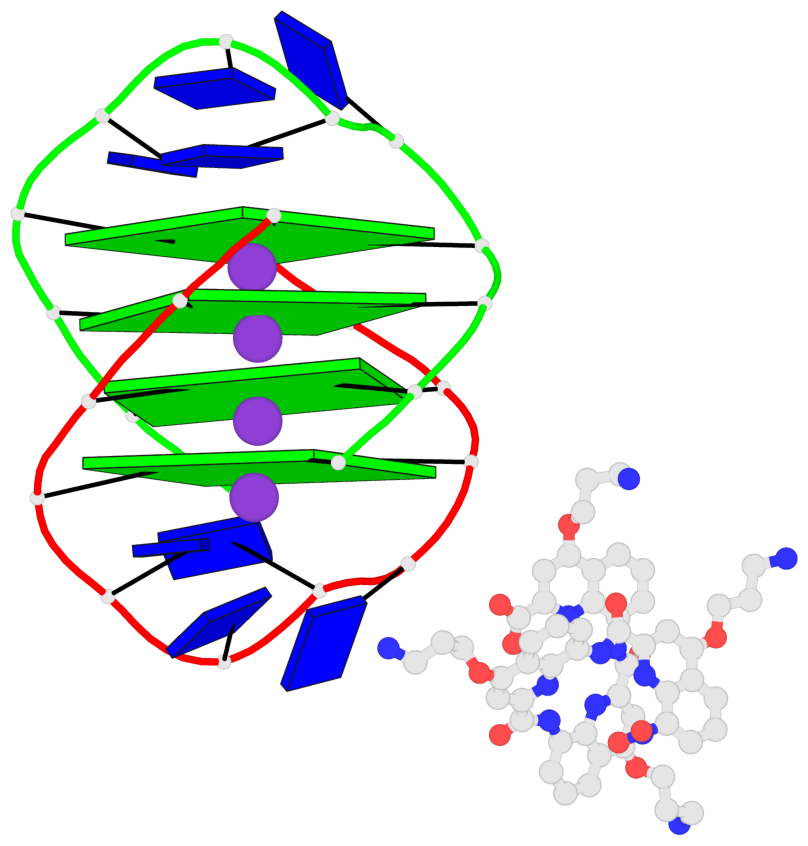
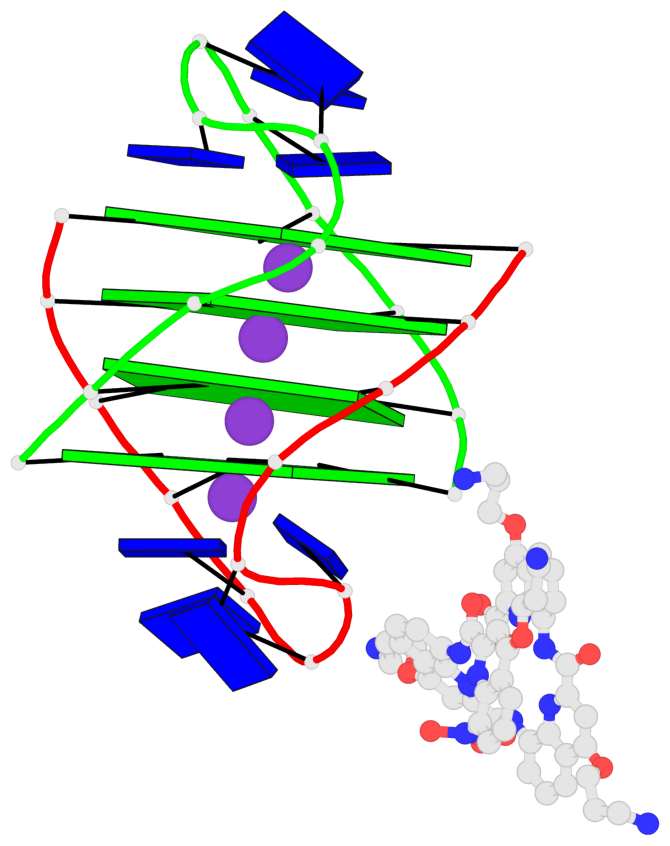
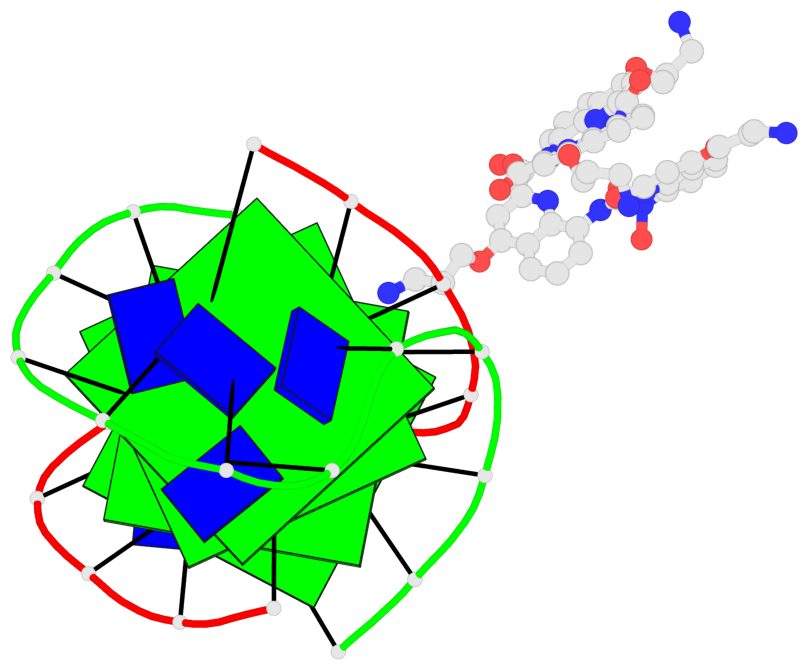
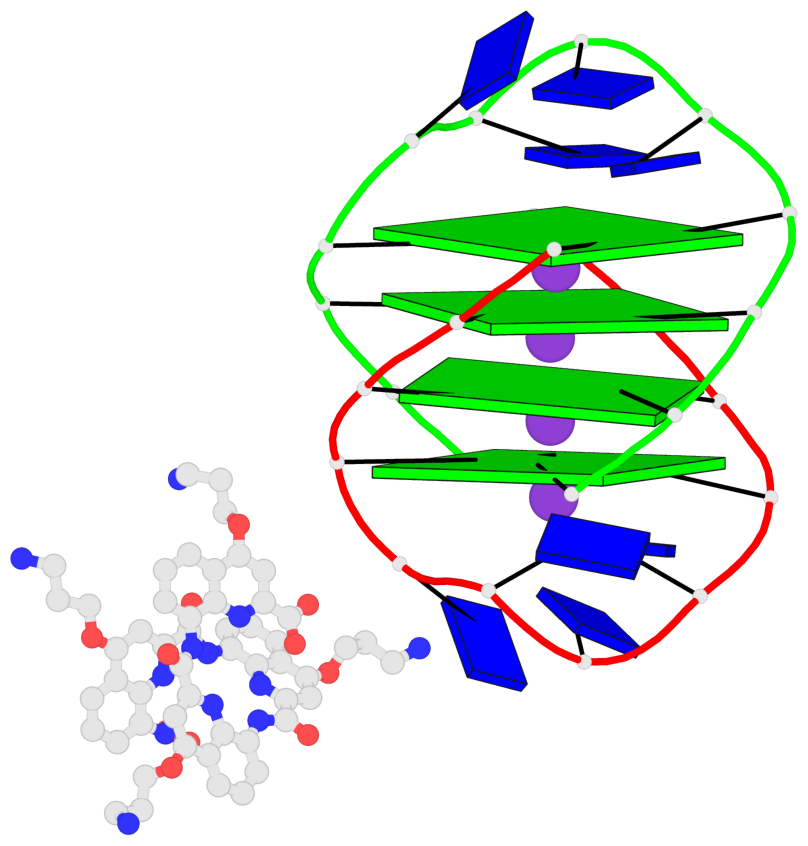
Base-block schematics in six views
List of 4 G-tetrads
1 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.160 type=other nts=4 GGGG A.DG1,B.DG4,A.DG12,B.DG9 2 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.137 type=planar nts=4 GGGG A.DG2,B.DG3,A.DG11,B.DG10 3 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.160 type=other nts=4 GGGG A.DG3,B.DG2,A.DG10,B.DG11 4 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.293 type=bowl nts=4 GGGG A.DG4,B.DG1,A.DG9,B.DG12
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.