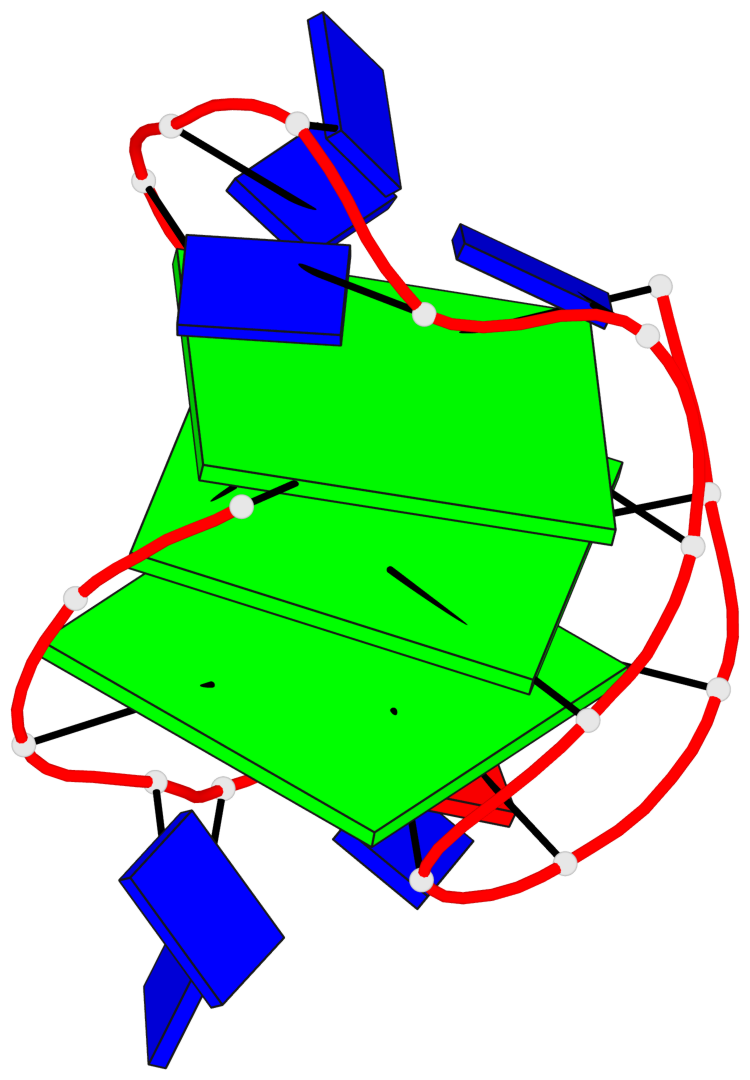
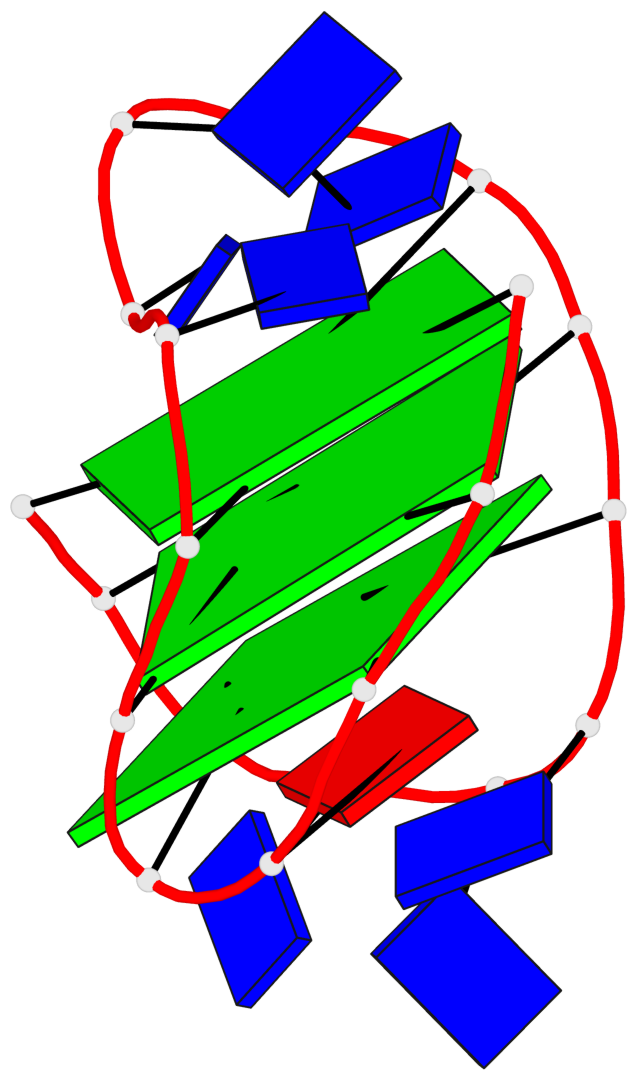
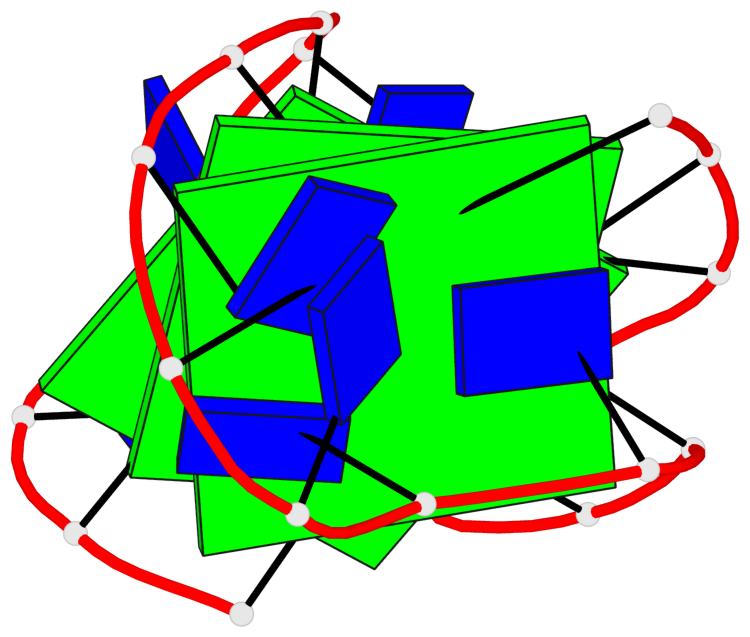
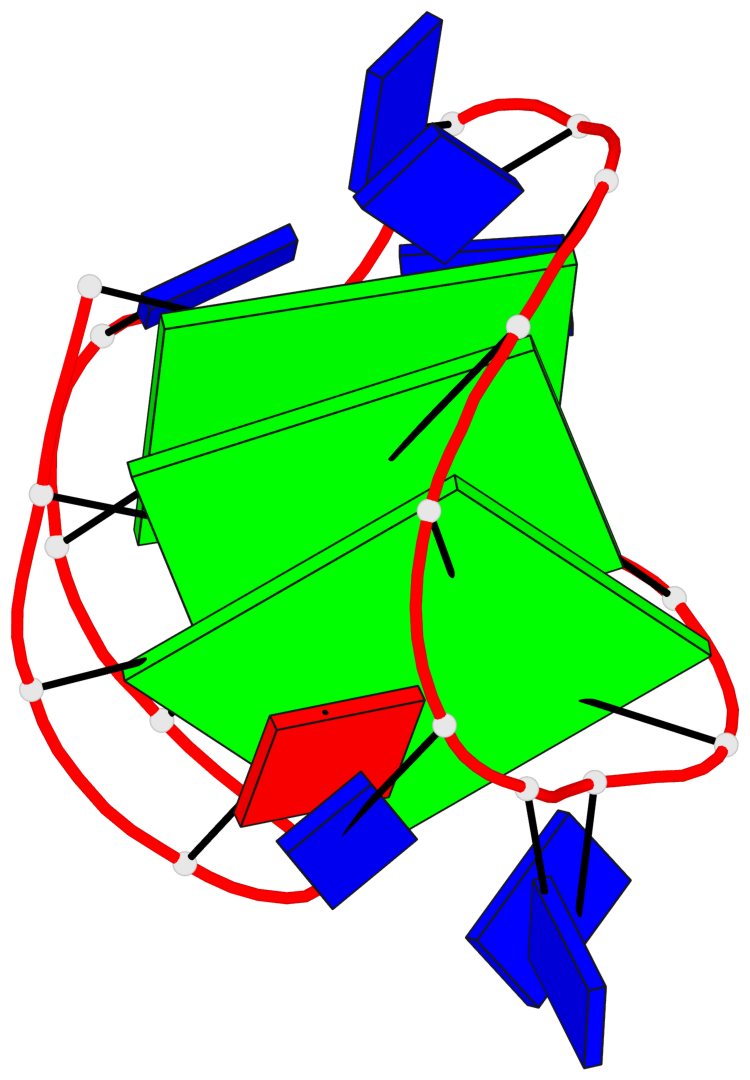
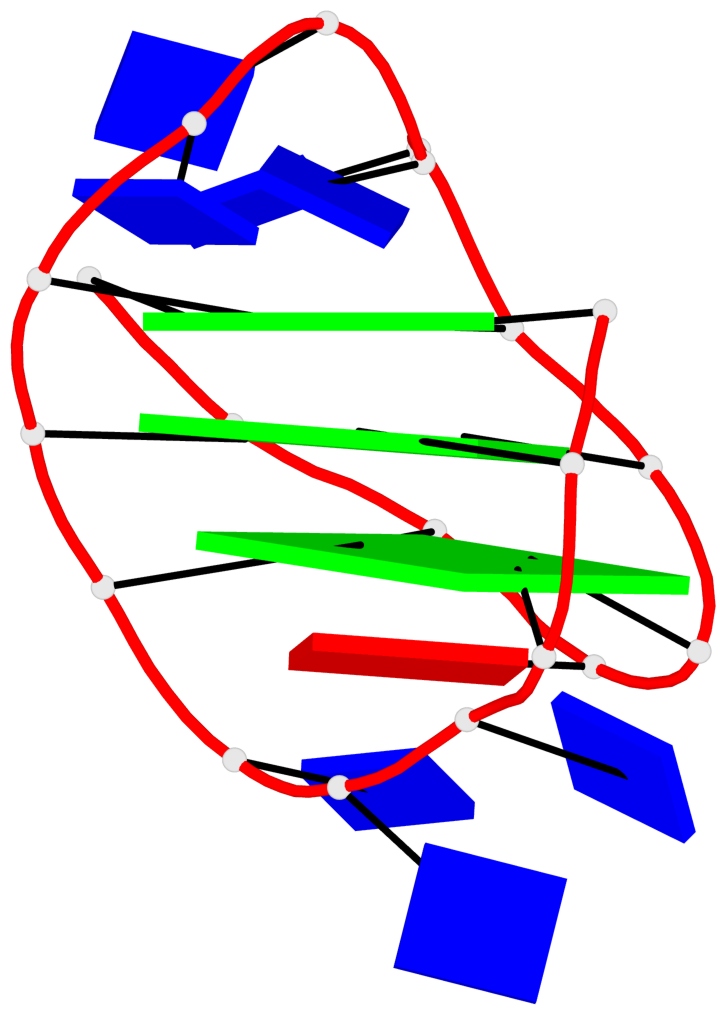
Detailed DSSR results for the G-quadruplex: PDB entry 5j05
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 5j05
- Class
- DNA
- Method
- NMR
- Summary
- Diy g-quadruplexes: solution structure of d(gggtttgggttttgggaggg) in sodium
- Reference
- Dvorkin SA, Karsisiotis AI, Webba da Silva M (2018): "Encoding canonical DNA quadruplex structure." Sci Adv, 4, eaat3007. doi: 10.1126/sciadv.aat3007.
- Abstract
- The main challenge in DNA quadruplex design is to encode a three-dimensional structure into the primary sequence, despite its multiple, repetitive guanine segments. We identify and detail structural elements describing all 14 feasible canonical quadruplex scaffolds and demonstrate their use in control of design. This work outlines a new roadmap for implementation of targeted design of quadruplexes for material, biotechnological, and therapeutic applications.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-LwD+Ln), basket(2+2), UDDU
Base-block schematics in six views
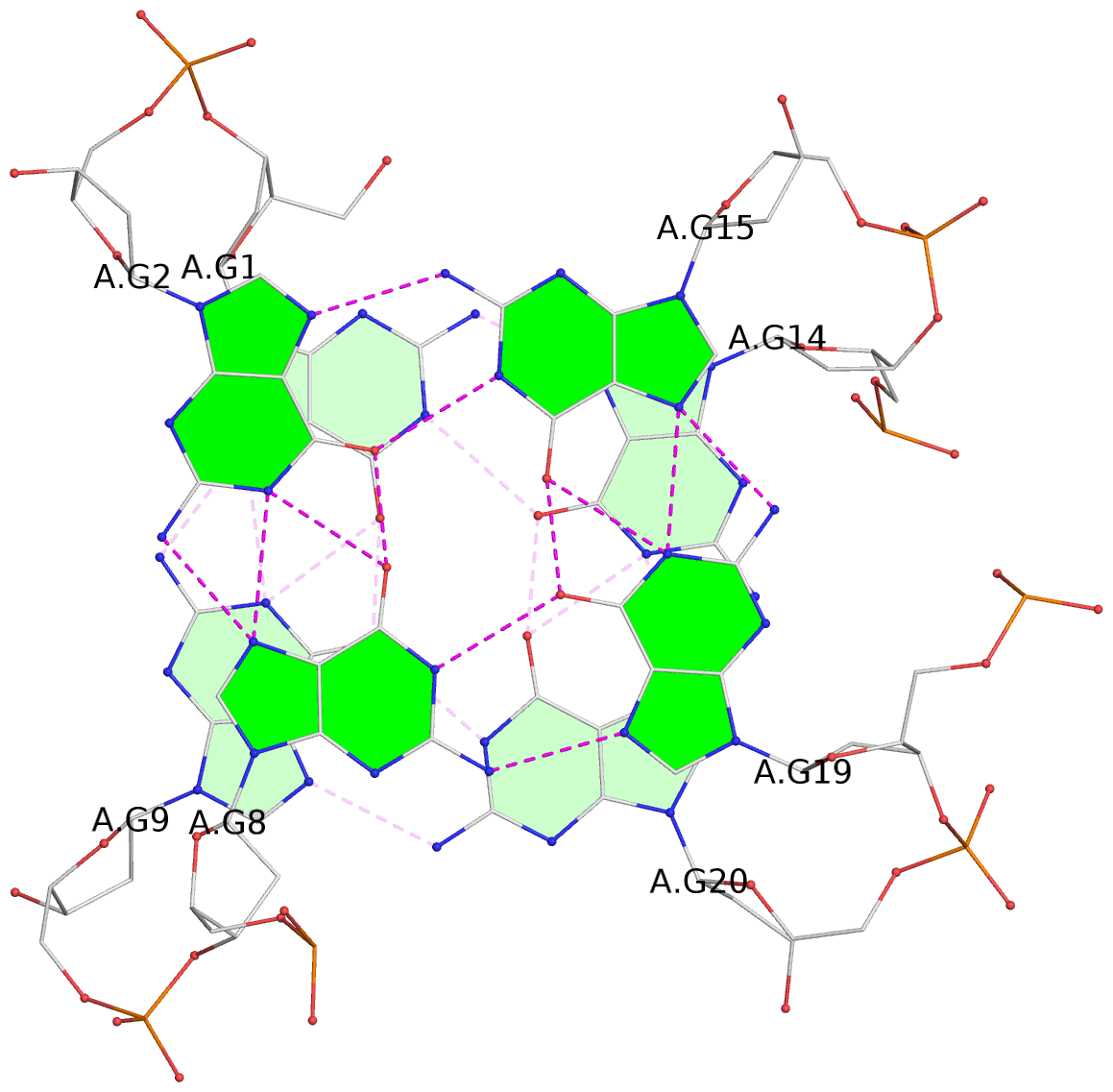
List of 3 G-tetrads
1 glyco-bond=s--s sugar=--.- groove=w-n- planarity=0.145 type=planar nts=4 GGGG A.DG1,A.DG9,A.DG20,A.DG14 2 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.151 type=planar nts=4 GGGG A.DG2,A.DG8,A.DG19,A.DG15 3 glyco-bond=s--s sugar=33-3 groove=w-n- planarity=0.255 type=other nts=4 GGGG A.DG3,A.DG7,A.DG18,A.DG16
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.