Detailed DSSR results for the G-quadruplex: PDB entry 5lig
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 5lig
- Class
- DNA
- Method
- NMR
- Summary
- G-quadruplex formed at the 5'-end of nheiii_1 element in human c-myc promoter bound to triangulenium based fluorescence probe daota-m2
- Reference
- Kotar A, Wang B, Shivalingam A, Gonzalez-Garcia J, Vilar R, Plavec J (2016): "NMR Structure of a Triangulenium-Based Long-Lived Fluorescence Probe Bound to a G-Quadruplex." Angew.Chem.Int.Ed.Engl., 55, 12508-12511. doi: 10.1002/anie.201606877.
- Abstract
- An NMR structural study of the interaction between a small-molecule optical probe (DAOTA-M2) and a G-quadruplex from the promoter region of the c-myc oncogene revealed that they interact at 1:2 binding stoichiometry. NMR-restrained structural calculations show that binding of DAOTA-M2 occurs mainly through π-π stacking between the polyaromatic core of the ligand and guanine residues of the outer G-quartets. Interestingly, the binding affinities of DAOTA-M2 differ by a factor of two for the outer G-quartets of the unimolecular parallel G-quadruplex under study. Unrestrained MD calculations indicate that DAOTA-M2 displays significant dynamic behavior when stacked on a G-quartet plane. These studies provide molecular guidelines for the design of triangulenium derivatives that can be used as optical probes for G-quadruplexes.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
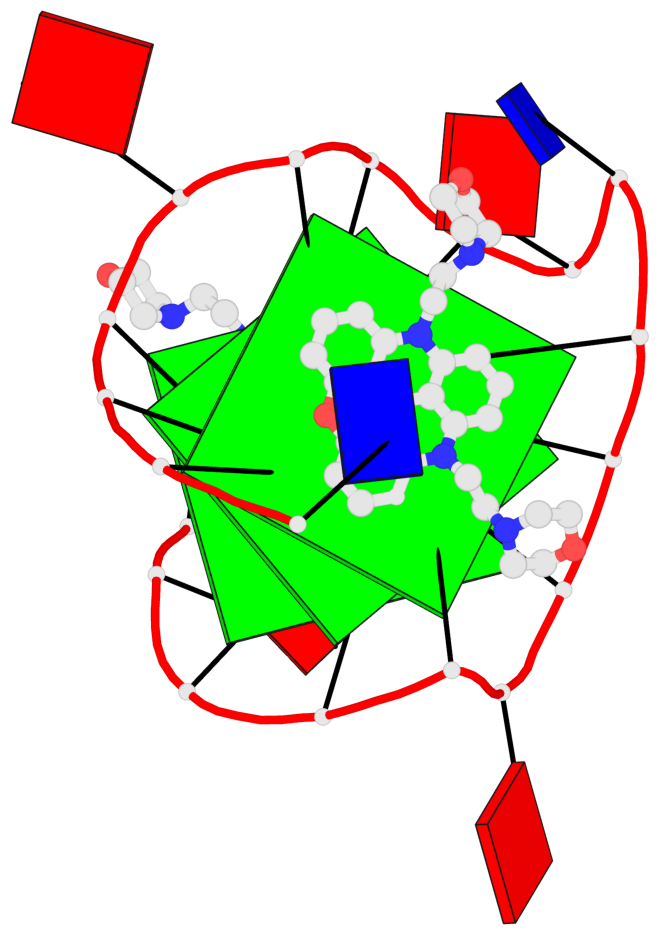
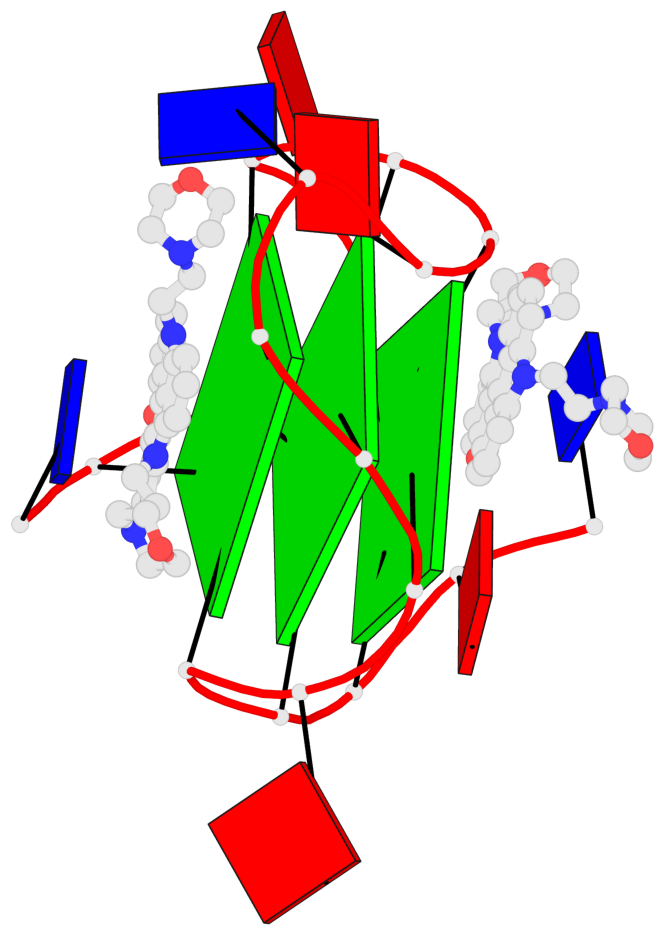
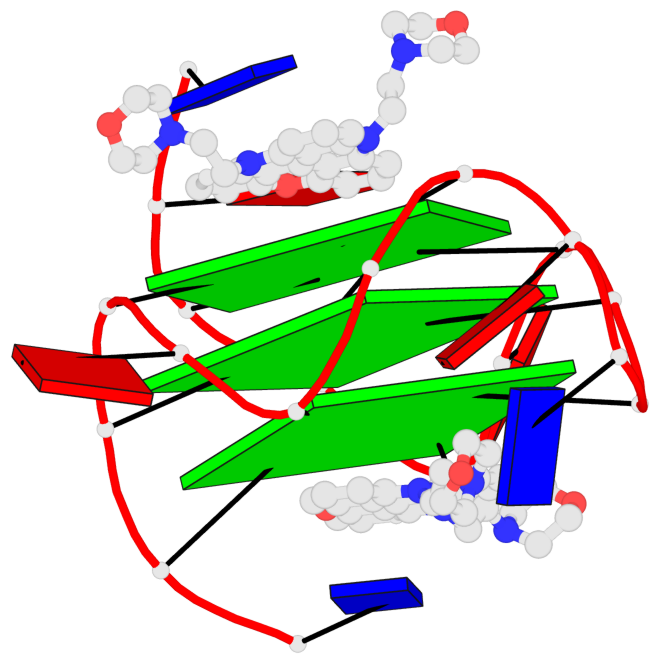
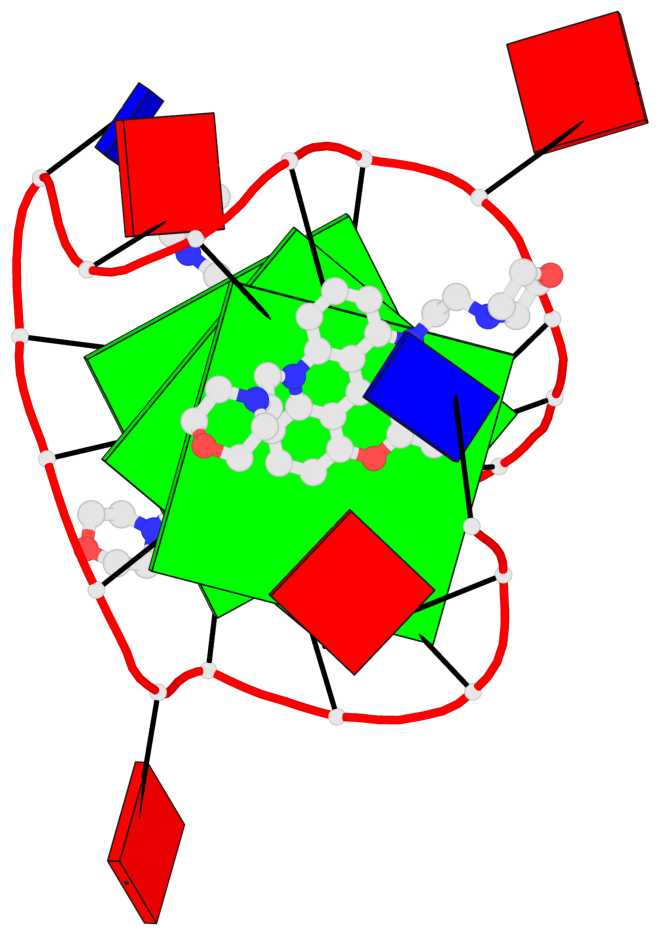
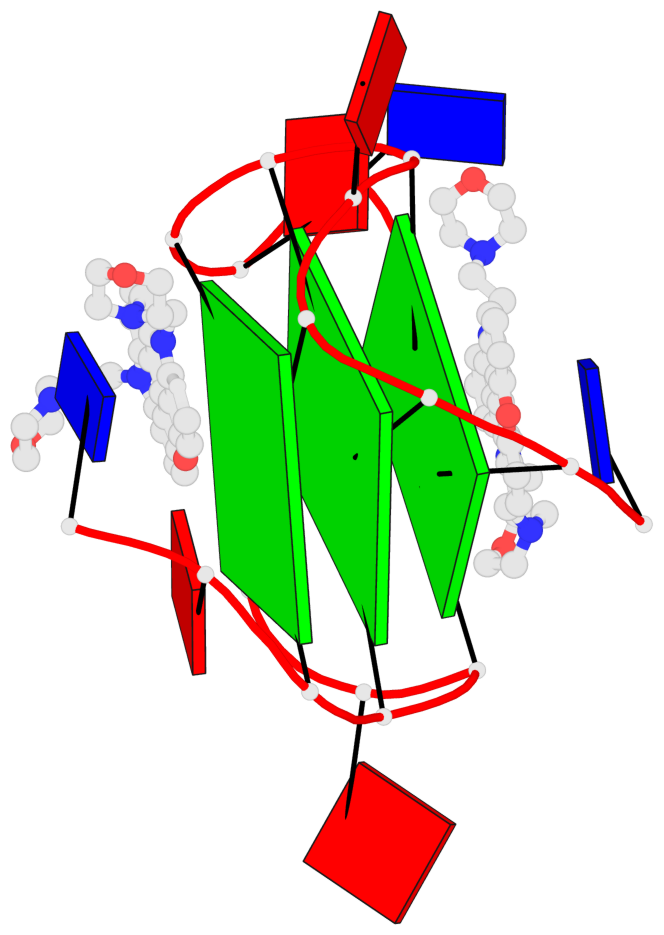
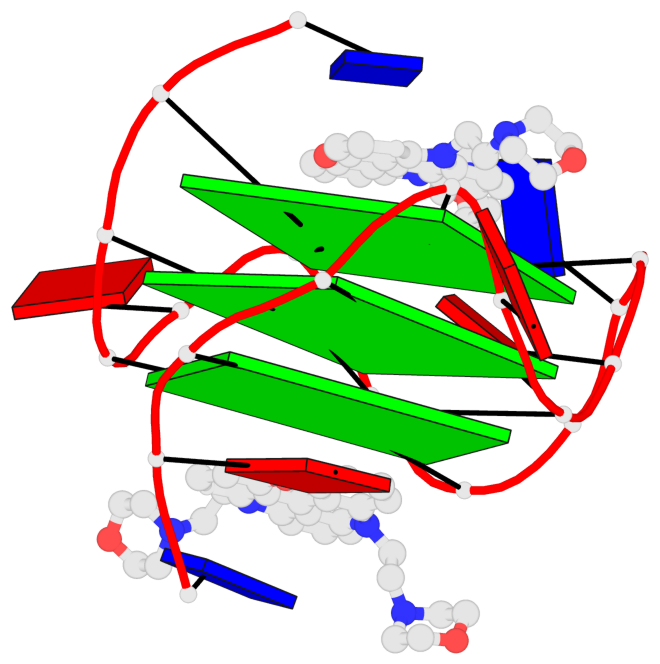
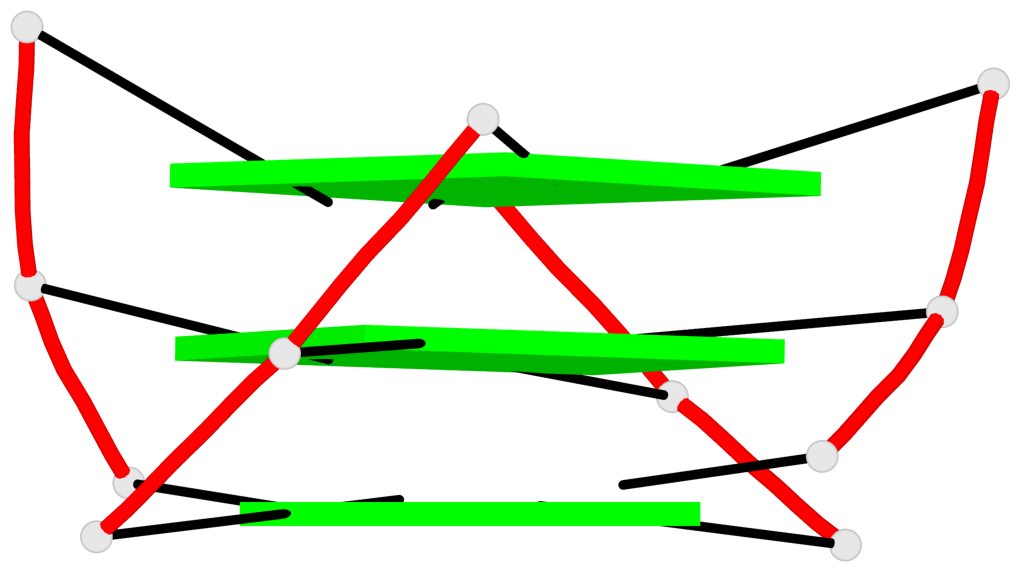
Base-block schematics in six views
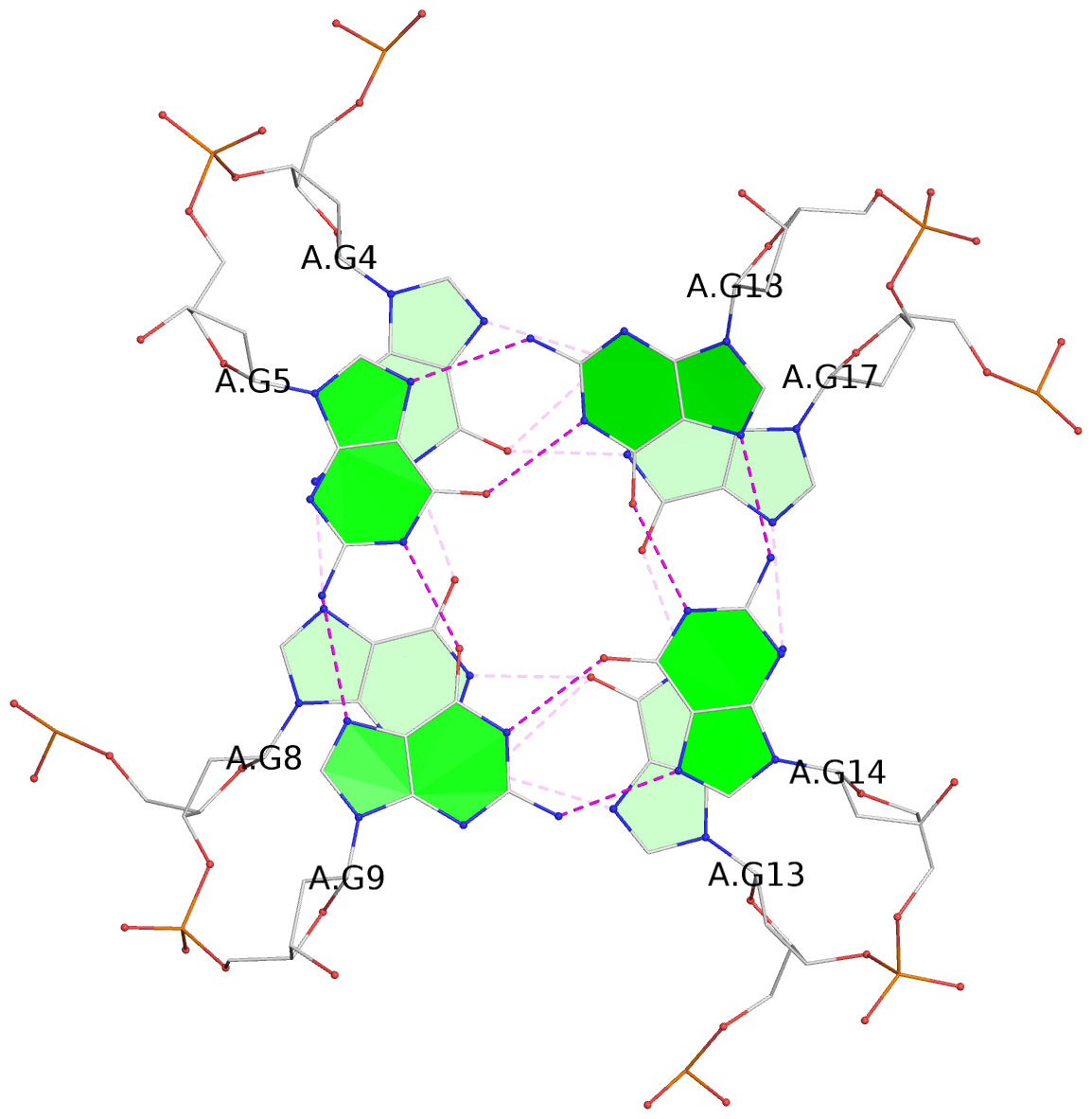
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.307 type=other nts=4 GGGG A.DG3,A.DG7,A.DG12,A.DG16 2 glyco-bond=---- sugar=---- groove=---- planarity=0.318 type=other nts=4 GGGG A.DG4,A.DG8,A.DG13,A.DG17 3 glyco-bond=---- sugar=---- groove=---- planarity=0.417 type=bowl nts=4 GGGG A.DG5,A.DG9,A.DG14,A.DG18
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.