Detailed DSSR results for the G-quadruplex: PDB entry 5lqh
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 5lqh
- Class
- DNA
- Method
- NMR
- Summary
- A two-quartet g-quadruplex formed by human telomere in kcl solution at ph 5.0
- Reference
- Galer P, Wang B, Sket P, Plavec J (2016): "Reversible pH Switch of Two-Quartet G-Quadruplexes Formed by Human Telomere." Angew.Chem.Int.Ed.Engl., 55, 1993-1997. doi: 10.1002/anie.201507569.
- Abstract
- A four-repeat human telomere DNA sequence without the 3'-end guanine, d[TAGGG(TTAGGG)2 TTAGG] (htel1-ΔG23) has been found to adopt two distinct two G-quartet antiparallel basket-type G-quadruplexes, TD and KDH(+) in presence of KCl. NMR, CD, and UV spectroscopy have demonstrated that topology of KDH(+) form is distinctive with unique protonated T18⋅A20(+) ⋅G5 base triple and other capping structural elements that provide novel insight into structural polymorphism and heterogeneity of G-quadruplexes in general. Specific stacking interactions amongst two G-quartets flanking base triples and base pairs in TD and KDH(+) forms are reflected in 10 K higher thermal stability of KDH(+) . Populations of TD and KDH(+) forms are controlled by pH. The (de)protonation of A20 is the key for pH driven structural transformation of htel1-ΔG23. Reversibility offers possibilities for its utilization as a conformational switch within different compartments of living cell enabling specific ligand and protein interactions.
- G4 notes
- 2 G-tetrads, 1 G4 helix, 1 G4 stem, 2(+LnD-Lw), basket(2+2), UUDD
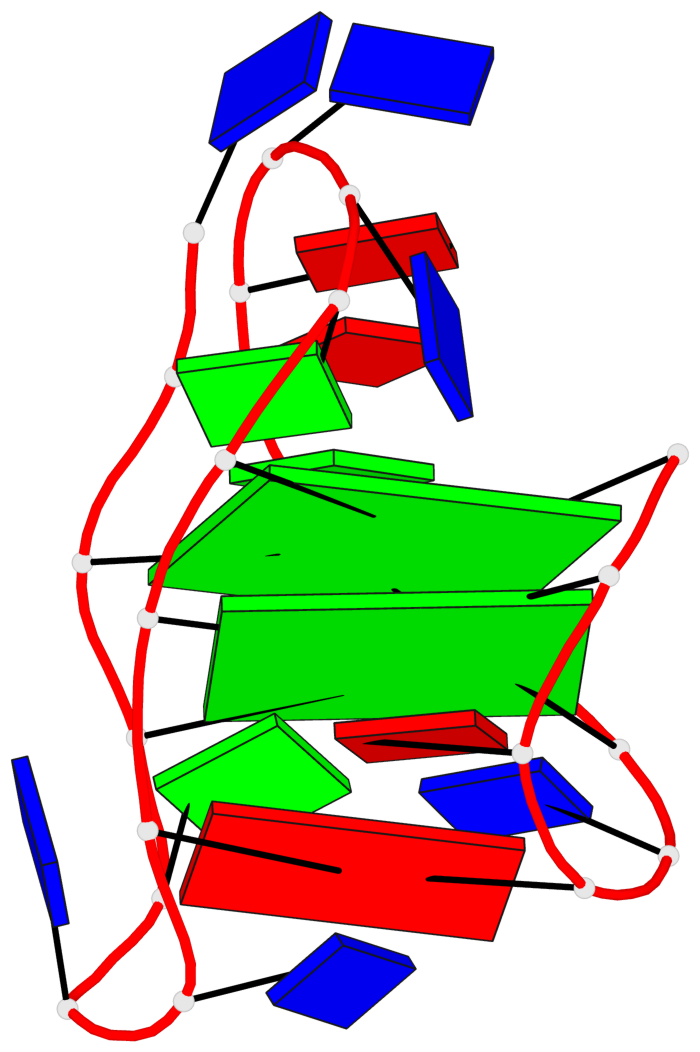
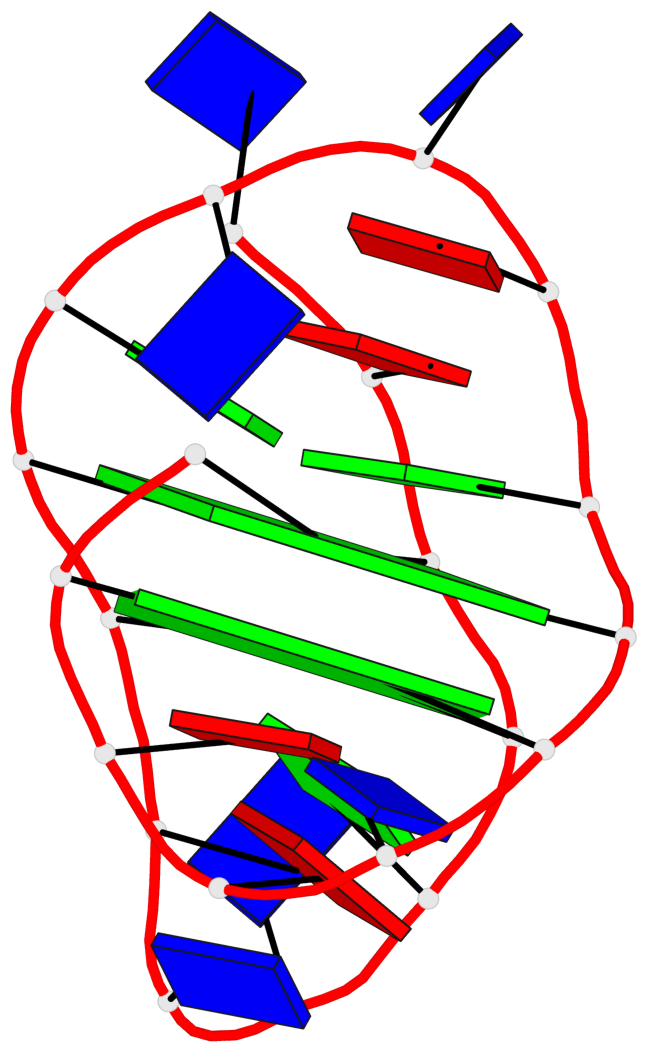
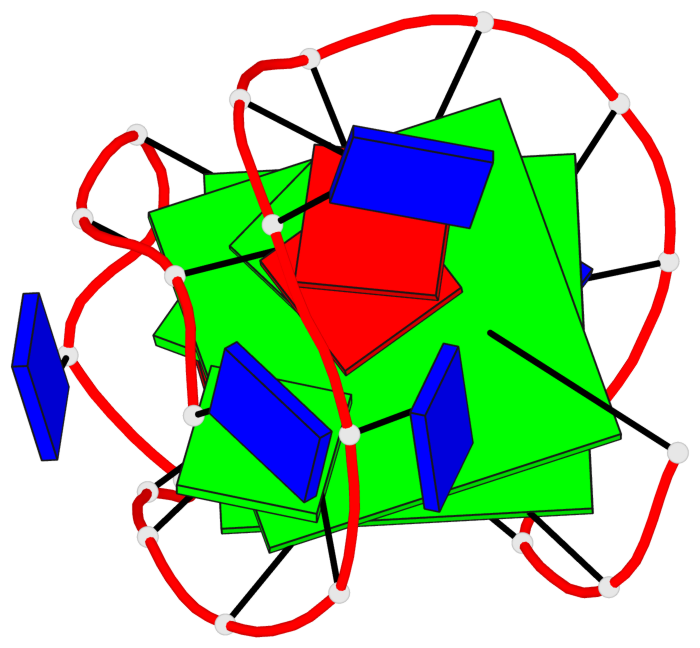
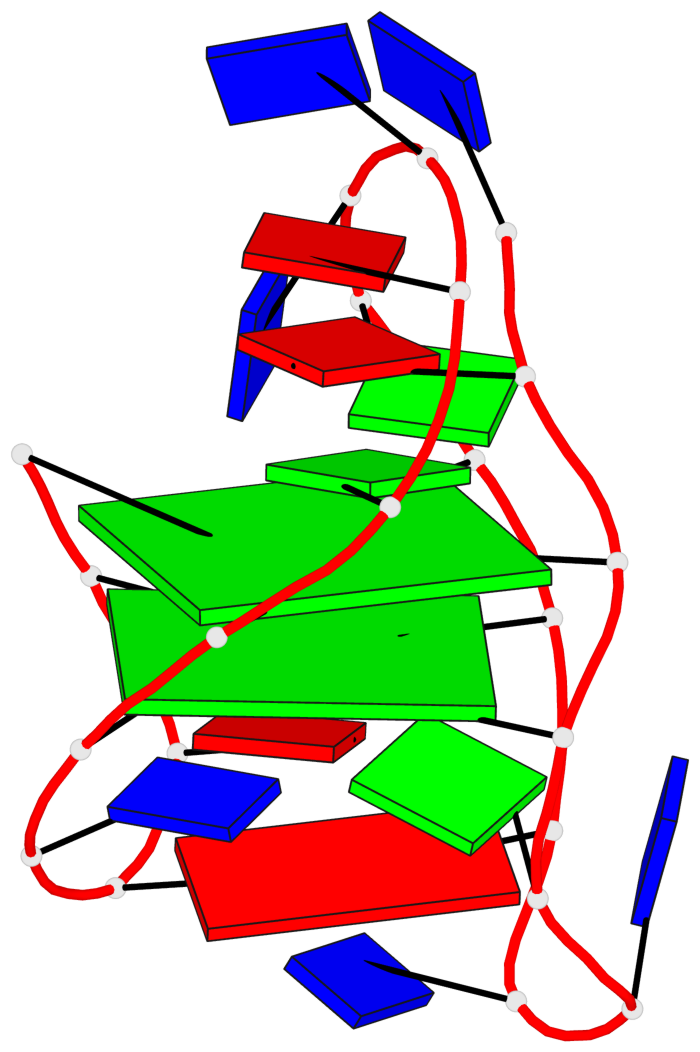
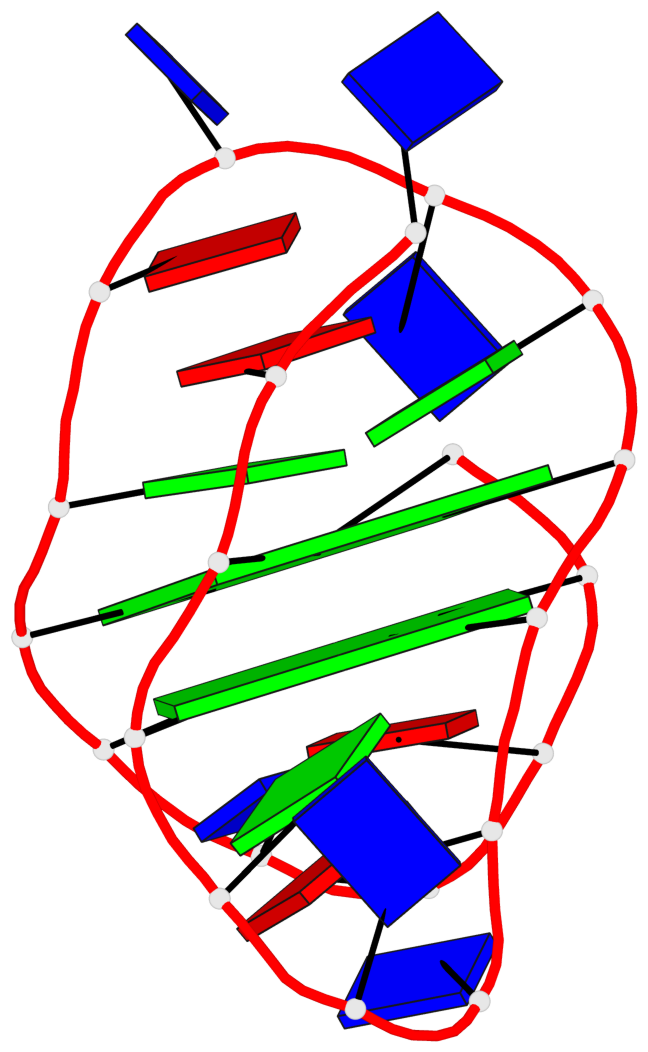
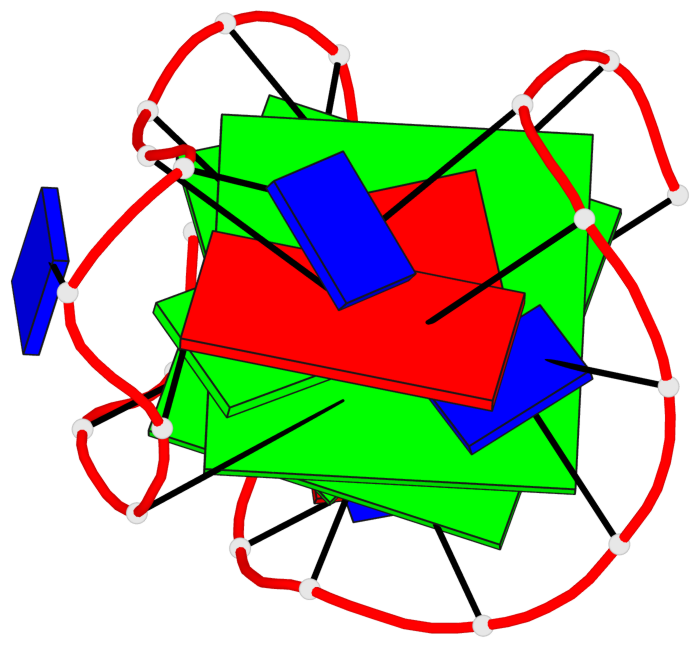
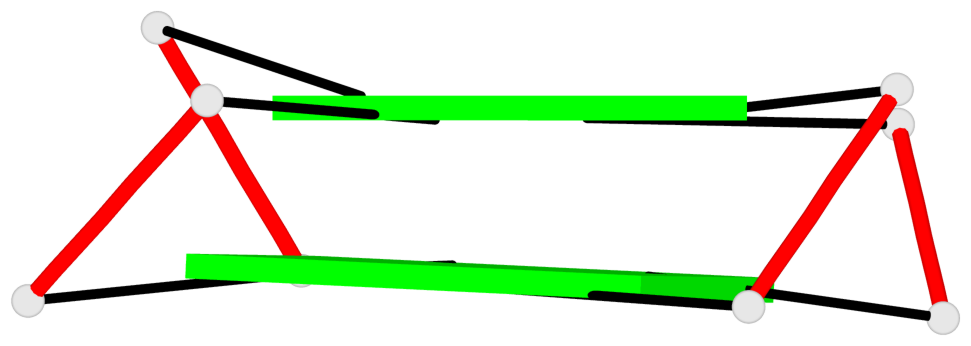
Base-block schematics in six views
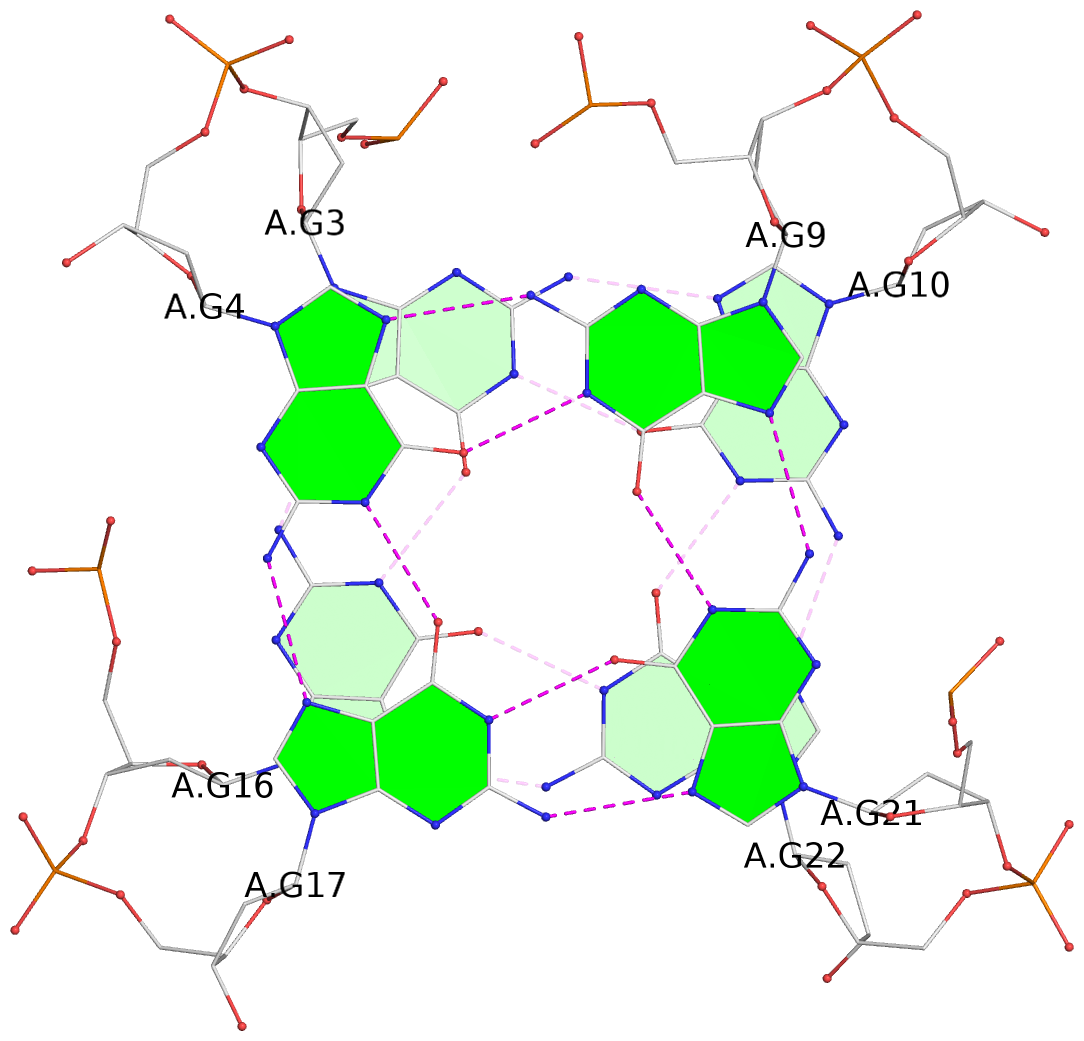
List of 2 G-tetrads
1 glyco-bond=ss-- sugar=---- groove=-w-n planarity=0.211 type=other nts=4 GGGG A.DG3,A.DG16,A.DG22,A.DG10 2 glyco-bond=--ss sugar=---- groove=-w-n planarity=0.154 type=planar nts=4 GGGG A.DG4,A.DG17,A.DG21,A.DG9
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.