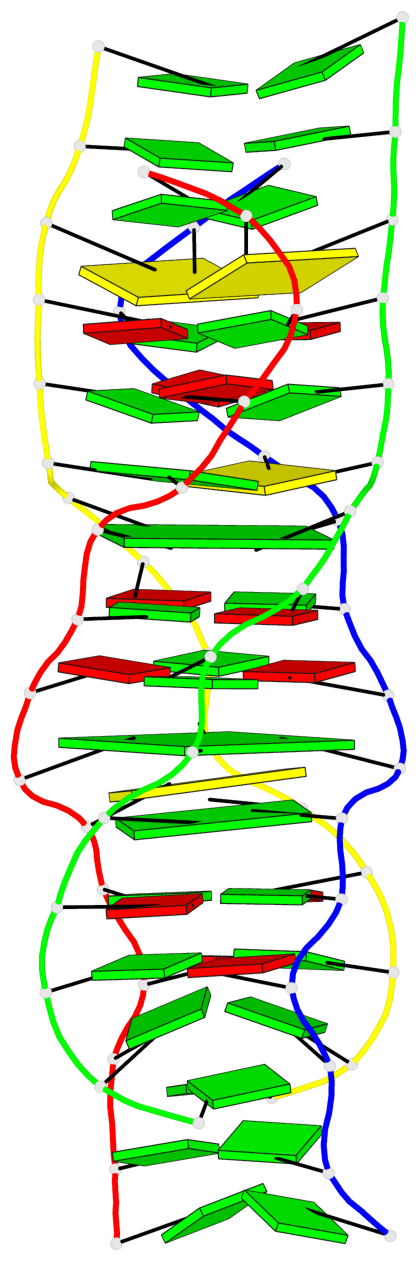
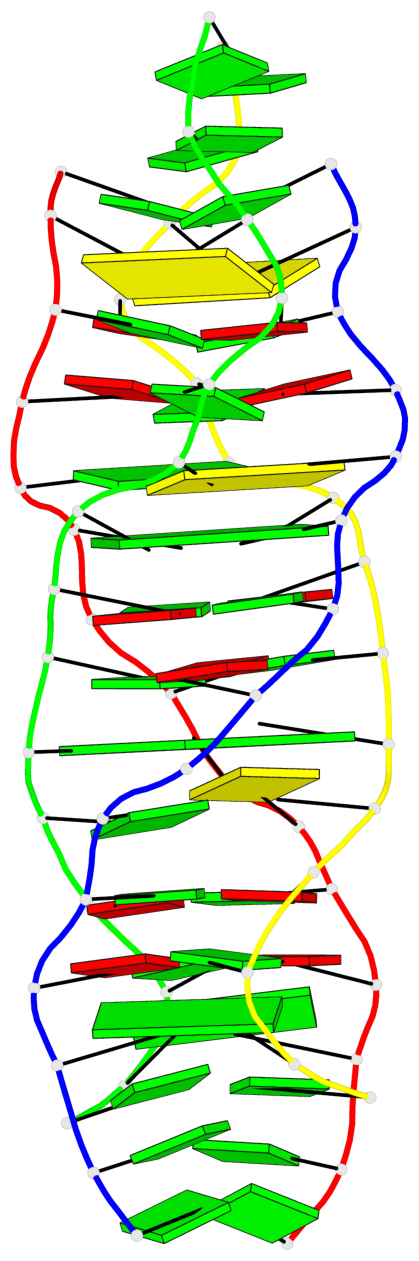
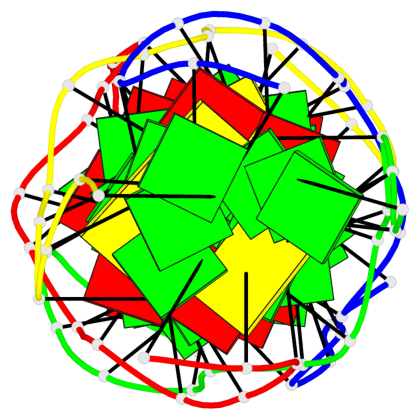
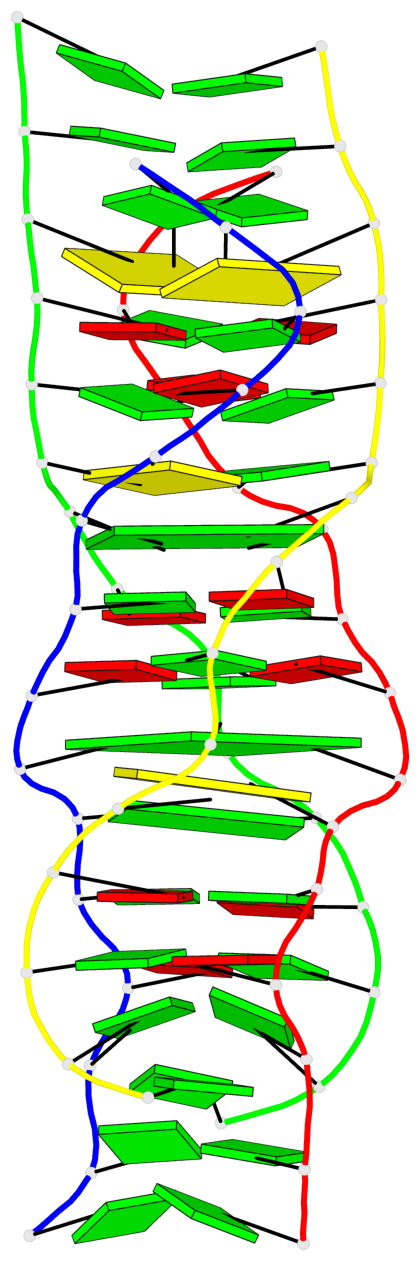
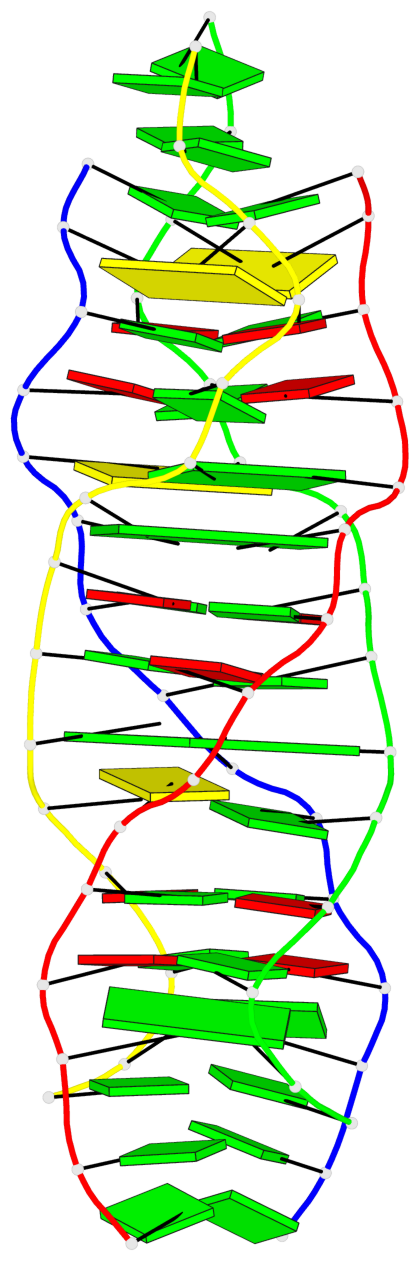
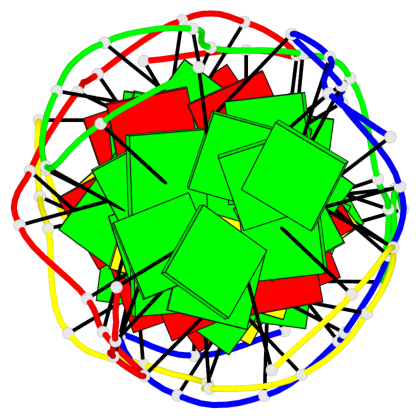
Detailed DSSR results for the G-quadruplex: PDB entry 5m2l
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 5m2l
- Class
- DNA
- Method
- NMR
- Summary
- Structure of DNA tetrameric agcga-quadruplex adopted by 15-mer d(gcgagggagcgaggg), vk34, oligonucleotide found in regulatory region of the plekhg3 human gene
- Reference
- Kocman V, Plavec J (2017): "Tetrahelical structural family adopted by AGCGA-rich regulatory DNA regions." Nat Commun, 8, 15355. doi: 10.1038/ncomms15355.
- Abstract
- Here we describe AGCGA-quadruplexes, an unexpected addition to the well-known tetrahelical families, G-quadruplexes and i-motifs, that have been a focus of intense research due to their potential biological impact in G- and C-rich DNA regions, respectively. High-resolution structures determined by solution-state nuclear magnetic resonance (NMR) spectroscopy demonstrate that AGCGA-quadruplexes comprise four 5'-AGCGA-3' tracts and are stabilized by G-A and G-C base pairs forming GAGA- and GCGC-quartets, respectively. Residues in the core of the structure are connected with edge-type loops. Sequences of alternating 5'-AGCGA-3' and 5'-GGG-3' repeats could be expected to form G-quadruplexes, but are shown herein to form AGCGA-quadruplexes instead. Unique structural features of AGCGA-quadruplexes together with lower sensitivity to cation and pH variation imply their potential biological relevance in regulatory regions of genes responsible for basic cellular processes that are related to neurological disorders, cancer and abnormalities in bone and cartilage development.
- G4 notes
- 2 G-tetrads
Base-block schematics in six views
List of 2 G-tetrads
1 glyco-bond=-s-s sugar=---. groove=wnwn planarity=0.457 type=other nts=4 GGGG A.DG6,D.DG9,C.DG6,B.DG9 2 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.467 type=other nts=4 GGGG A.DG9,D.DG6,C.DG9,B.DG6