Detailed DSSR results for the G-quadruplex: PDB entry 5mcr
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 5mcr
- Class
- DNA
- Method
- NMR
- Summary
- Quadruplex with flipped tetrad formed by an artificial sequence
- Reference
- Dickerhoff J, Haase L, Langel W, Weisz K (2017): "Tracing Effects of Fluorine Substitutions on G-Quadruplex Conformational Changes." ACS Chem. Biol., 12, 1308-1315. doi: 10.1021/acschembio.6b01096.
- Abstract
- A human telomere sequence that folds into an intramolecular (3 + 1)-hybrid G-quadruplex was modified by the incorporation of 2'-fluoro-2'-deoxyriboguanosines (FG) into syn positions of its outer tetrad. A circular dichroism and NMR spectral analysis reveals a nearly quantitative switch of the G-tetrad polarity with concerted syn↔anti transitions of all four G residues. These observations follow findings on a FG-substituted (3 + 1)-hybrid quadruplex with a different fold, suggesting a more general propensity of hybrid-type quadruplexes to undergo a tetrad polarity reversal. Two out of the three FG analogs in both modified quadruplexes adopt an S-type sugar pucker, challenging a sole contribution of N-type sugars in enforcing an anti glycosidic torsion angle associated with the tetrad flip. NMR restrained three-dimensional structures of the two substituted quadruplexes reveal a largely conserved overall fold but significant rearrangements of the overhang and loop nucleotides capping the flipped tetrad. Sugar pucker preferences of the FG analogs may be rationalized by different orientations of the fluorine atom and its resistance to be positioned within the narrow groove with its highly negative electrostatic potential and spine of water molecules.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-PD+Lw), U3D(3+1), UUUD
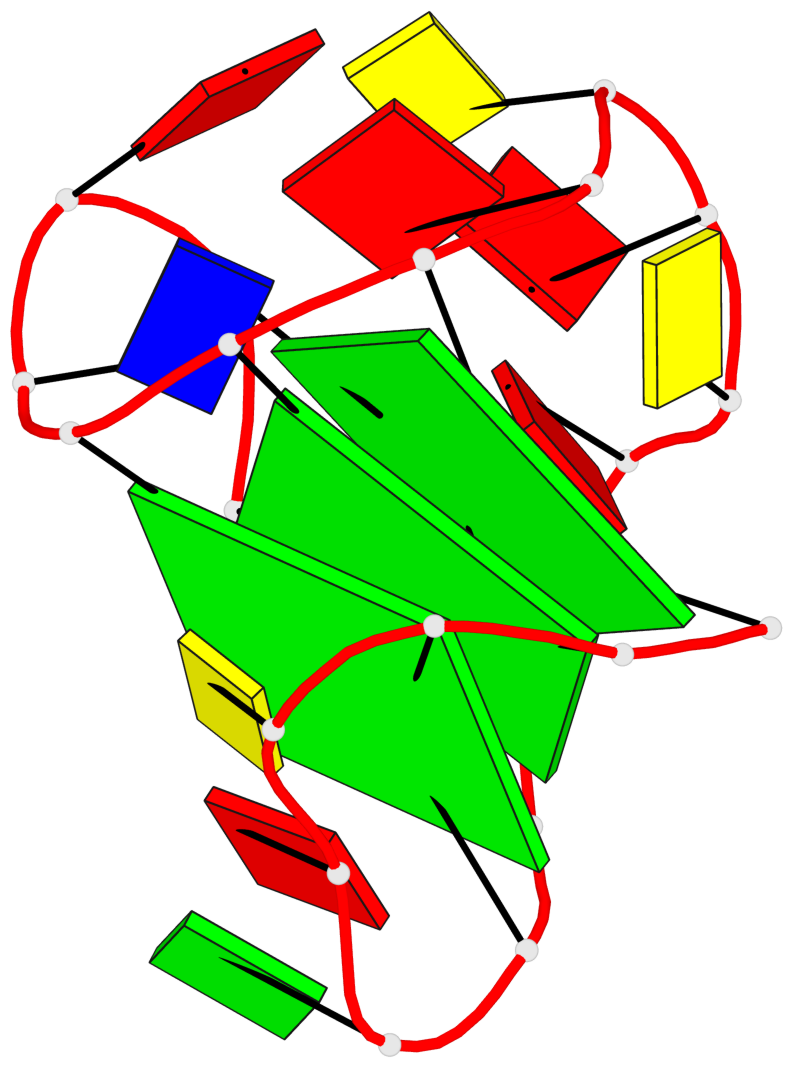
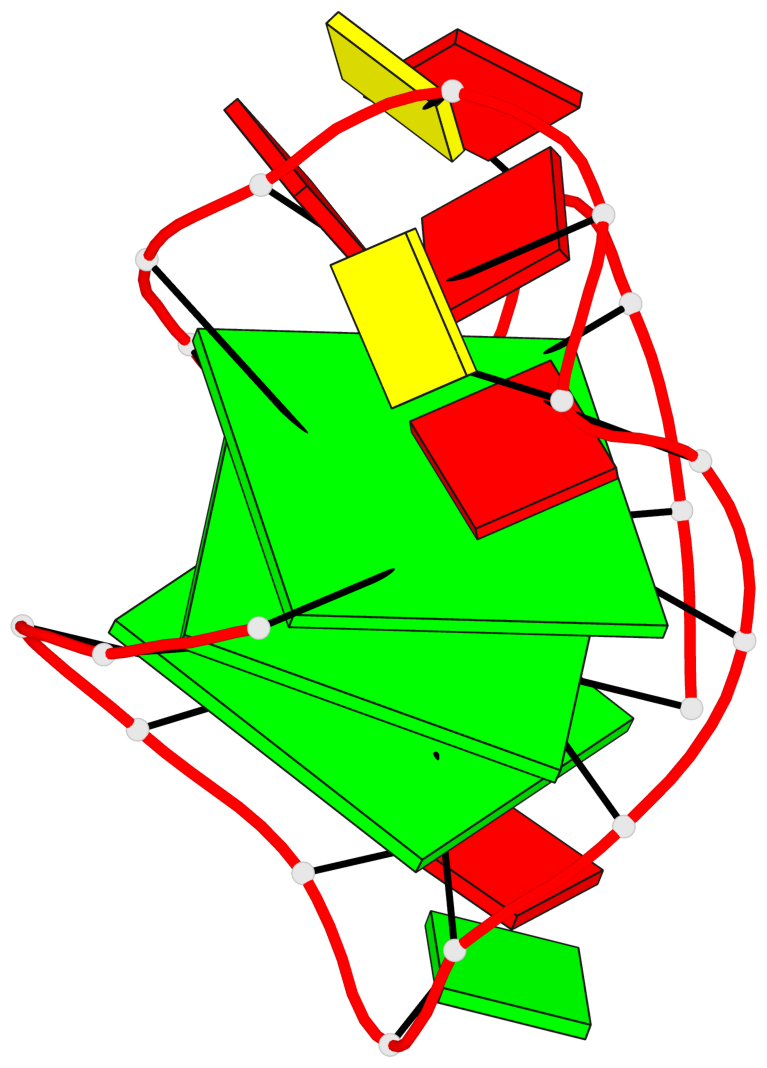
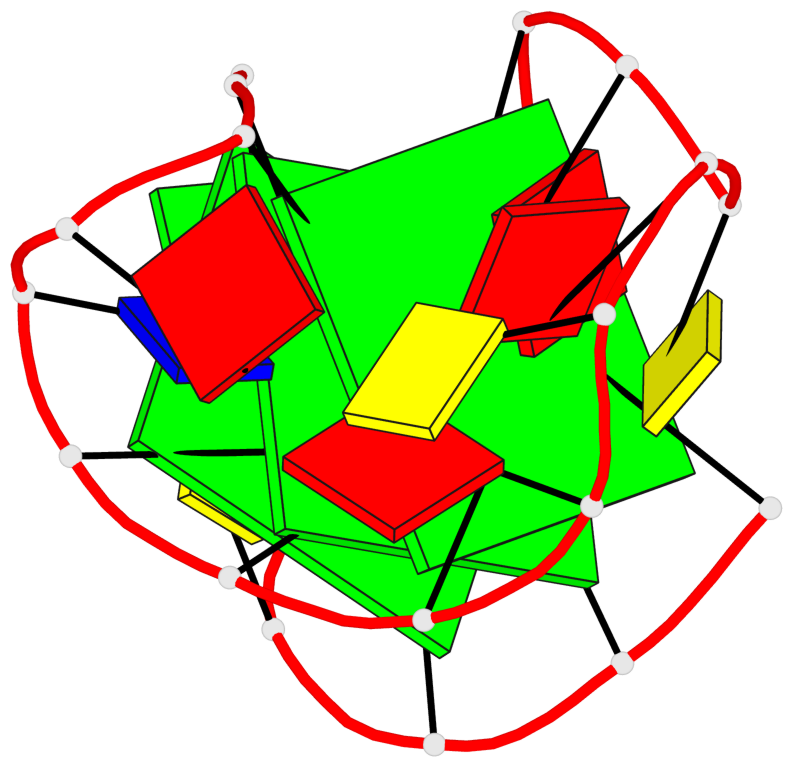
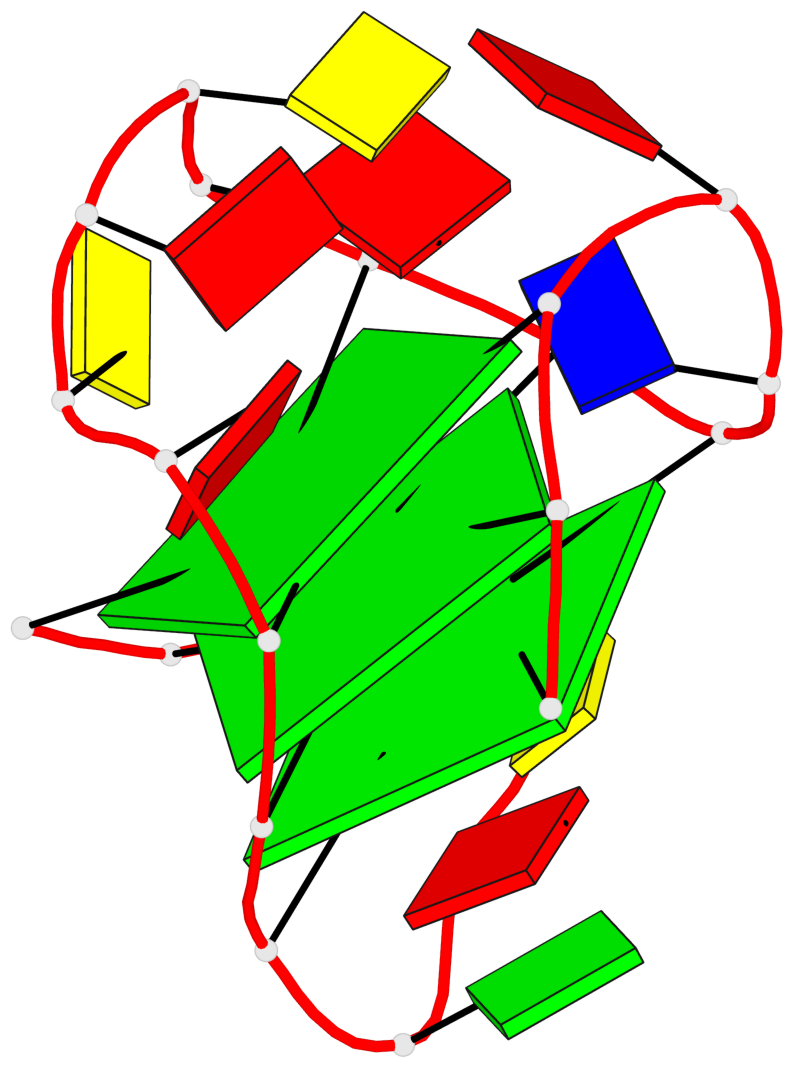
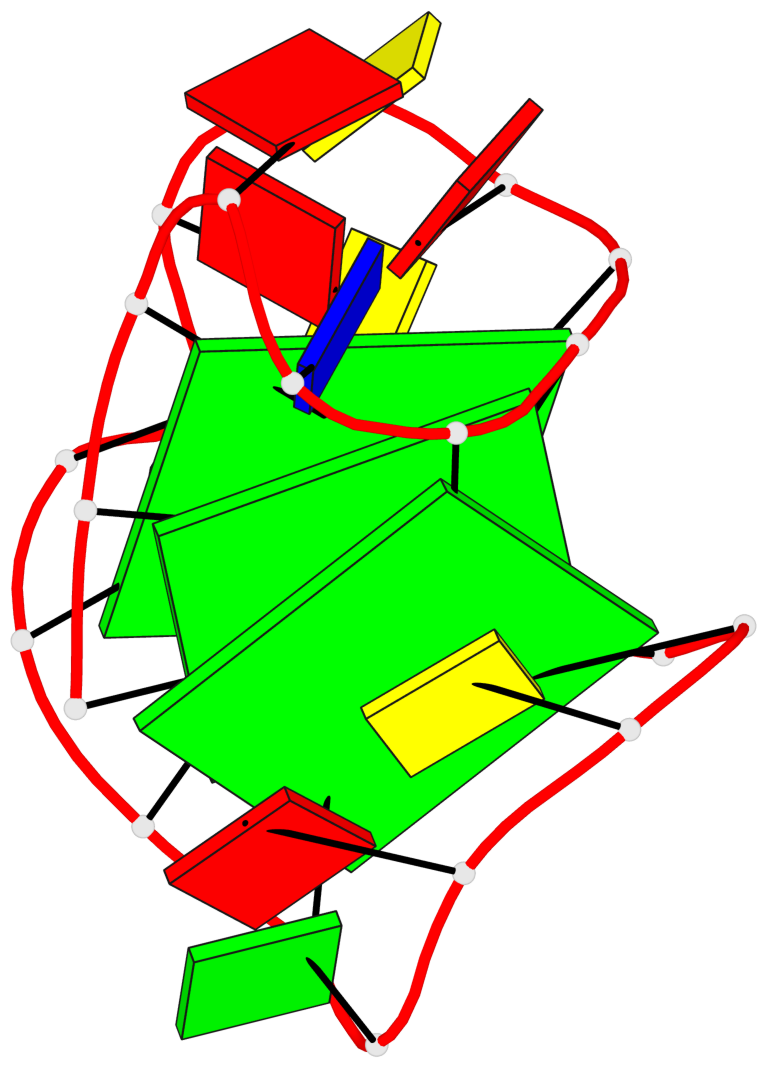
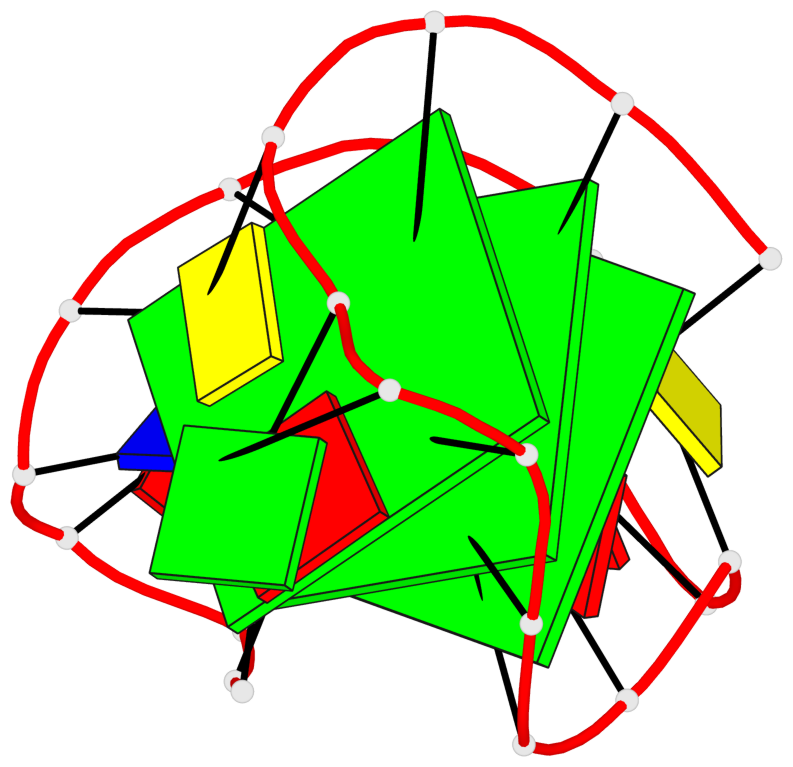
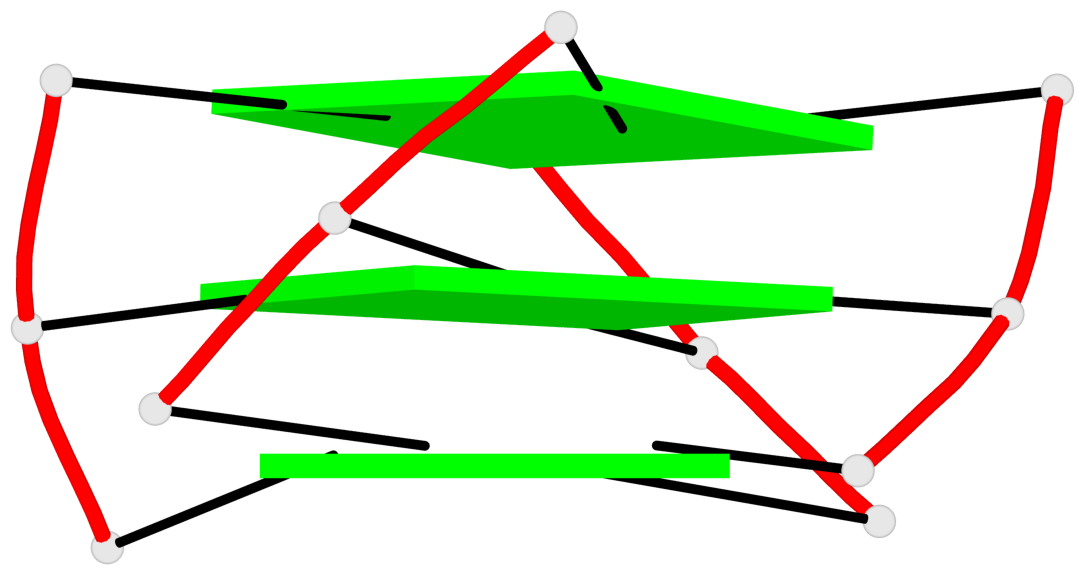
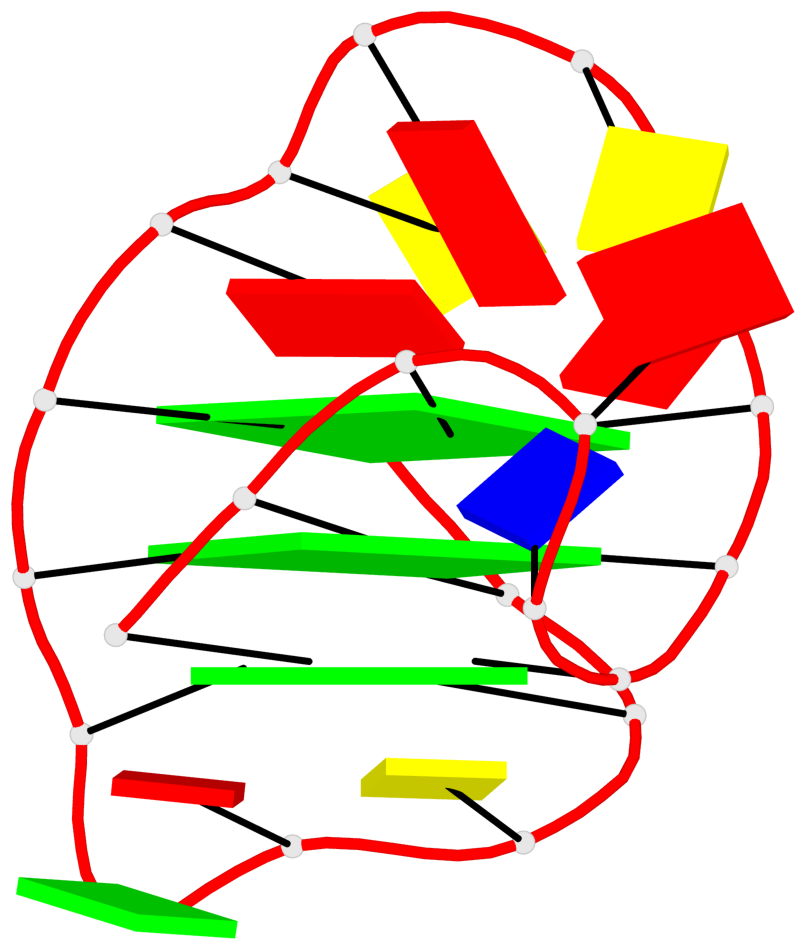
Base-block schematics in six views
List of 3 G-tetrads
1 glyco-bond=---s sugar=3--. groove=--wn planarity=0.292 type=other nts=4 gggG A.GF0/1,A.GF2/6,A.GF2/20,A.DG16 2 glyco-bond=---s sugar=3--- groove=--wn planarity=0.184 type=other nts=4 GGGG A.DG2,A.DG7,A.DG21,A.DG15 3 glyco-bond=---s sugar=---- groove=--wn planarity=0.244 type=other nts=4 GGGG A.DG3,A.DG8,A.DG22,A.DG14
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.