Detailed DSSR results for the G-quadruplex: PDB entry 5nyt
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 5nyt
- Class
- DNA
- Method
- NMR
- Summary
- M2 g-quadruplex 20 wt% ethylene glycol
- Reference
- Trajkovski M, Endoh T, Tateishi-Karimata H, Ohyama T, Tanaka S, Plavec J, Sugimoto N (2018): "Pursuing origins of (poly)ethylene glycol-induced G-quadruplex structural modulations." Nucleic Acids Res., 46, 4301-4315. doi: 10.1093/nar/gky250.
- Abstract
- Molecular crowding conditions provided by high concentration of cosolutes are utilized for characterization of biomolecules in cell-mimicking environment and development of drug-delivery systems. In this context, (poly)ethylene glycols are often used for studying non-canonical DNA structures termed G-quadruplexes, which came into focus by emerging structural biology findings and new therapeutic drug design approaches. Recently, several reports were made arguing against using (poly)ethylene glycols in role of molecular crowding agents due to their direct impact on DNA G-quadruplex stability and topology. However, the available data on structural details underlying DNA interaction is very scarce and thus limits in-depth comprehension. Herein, structural and thermodynamic analyses were strategically combined to assess G-quadruplex-cosolute interactions and address previously reported variances regarding the driving forces of G-rich DNA structural transformations under molecular crowding conditions. With the use of complementary (CD, NMR and UV) spectroscopic methods and model approach we characterized DNA G-quadruplex in the presence of the smallest and one of the largest typically used (poly)ethylene glycols. Dehydration effect is the key contributor to ethylene-glycol-induced increased stability of the G-quadruplex, which is in the case of the large cosolute mainly guided by the subtle direct interactions between PEG 8000 and the outer G-quartet regions.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
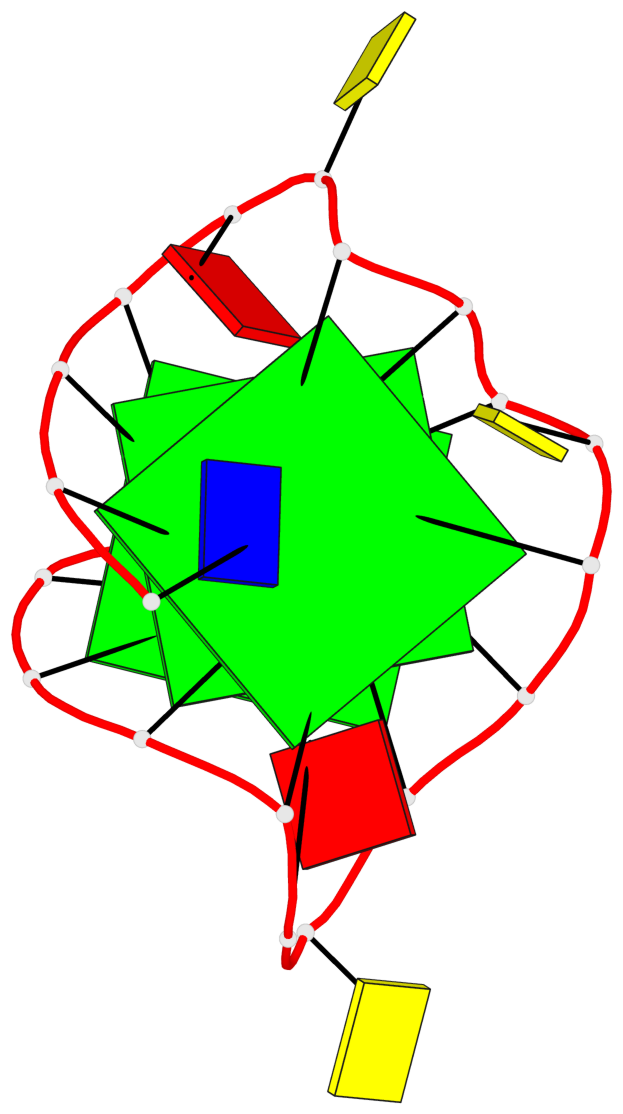
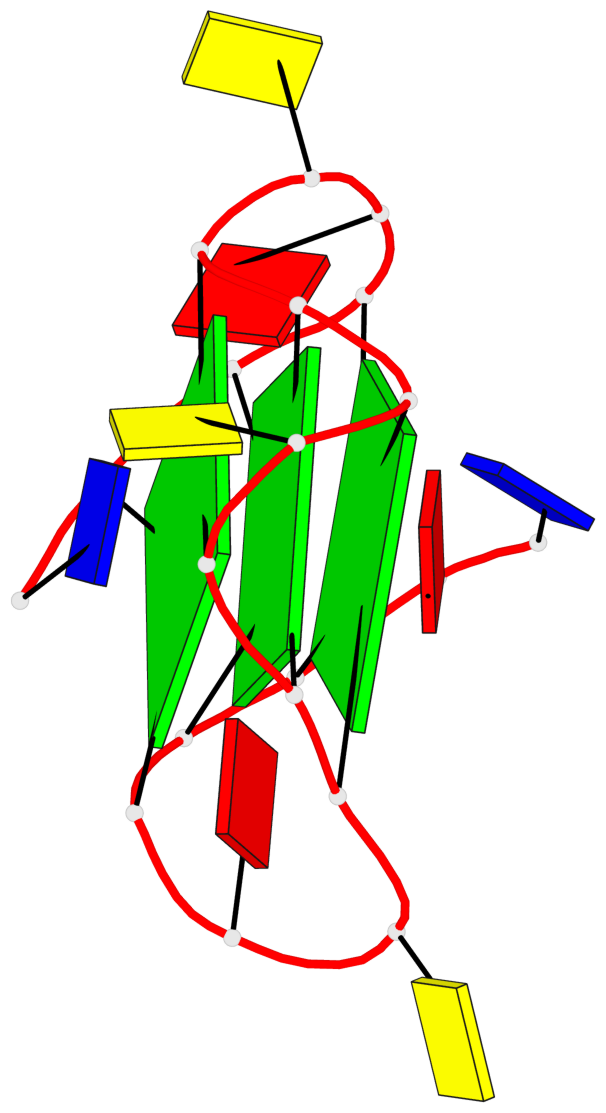
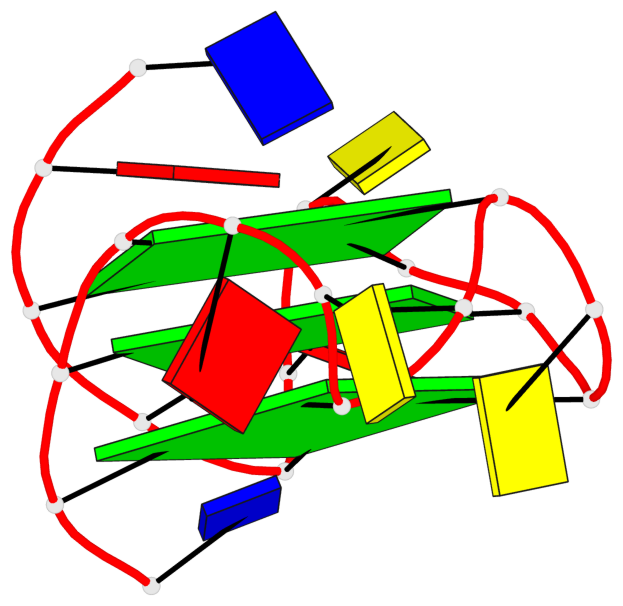
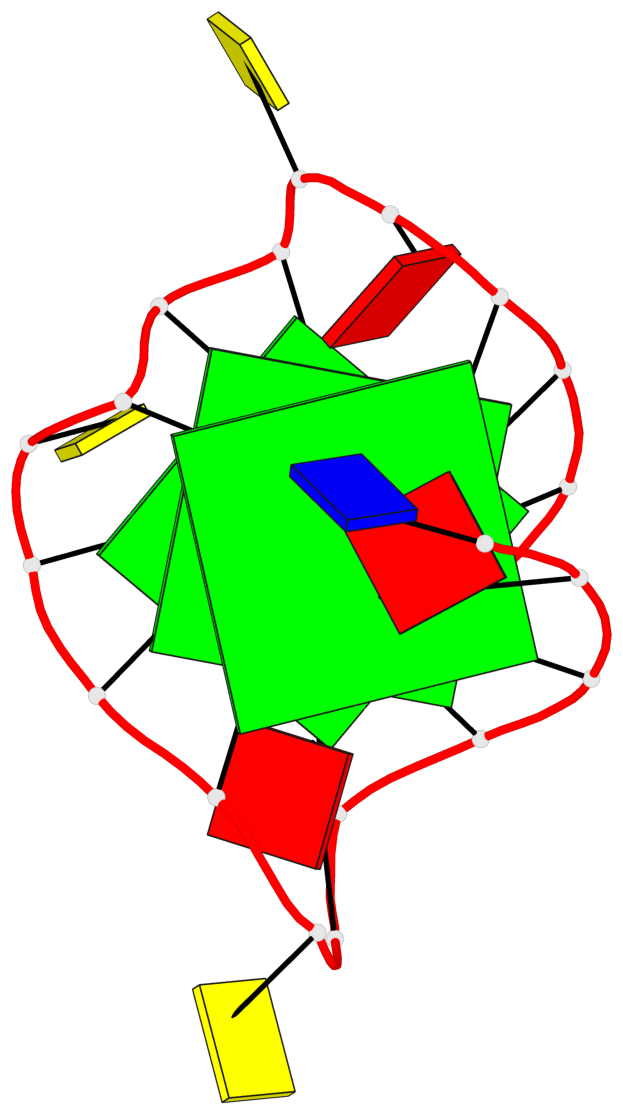
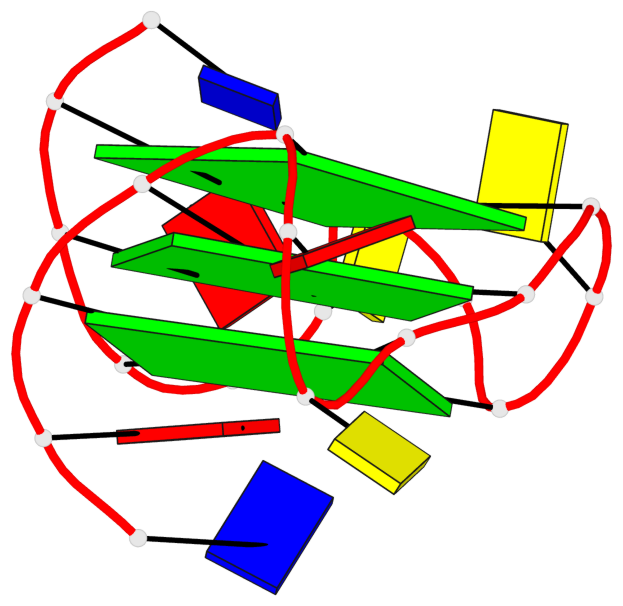
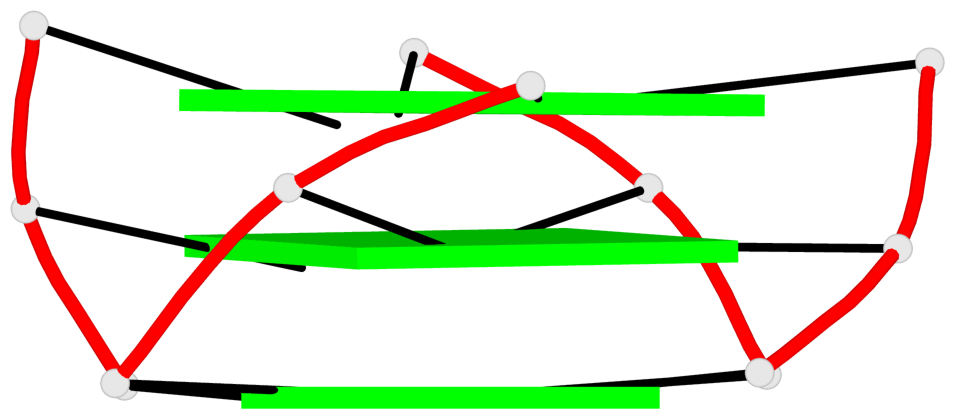
Base-block schematics in six views
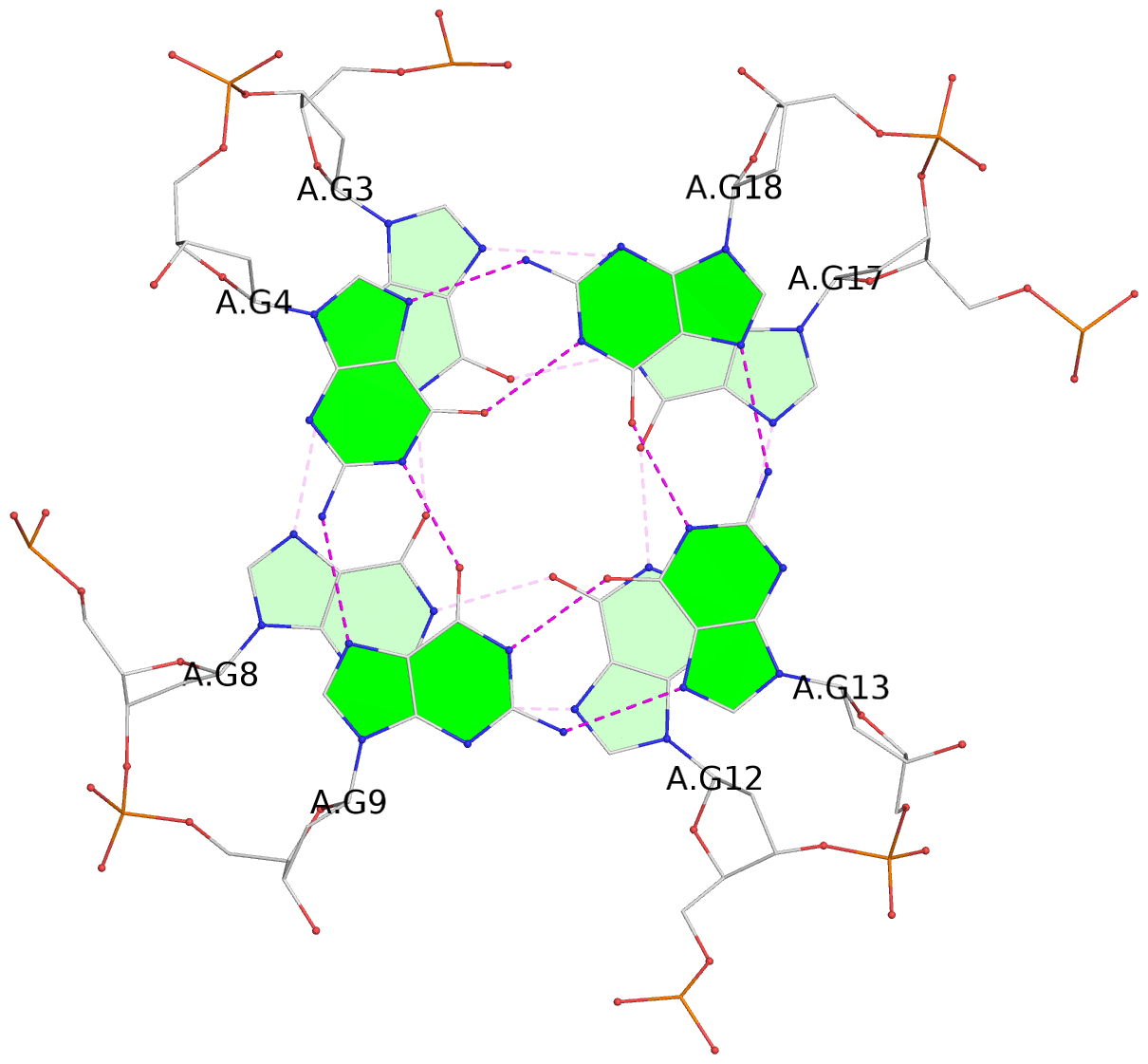
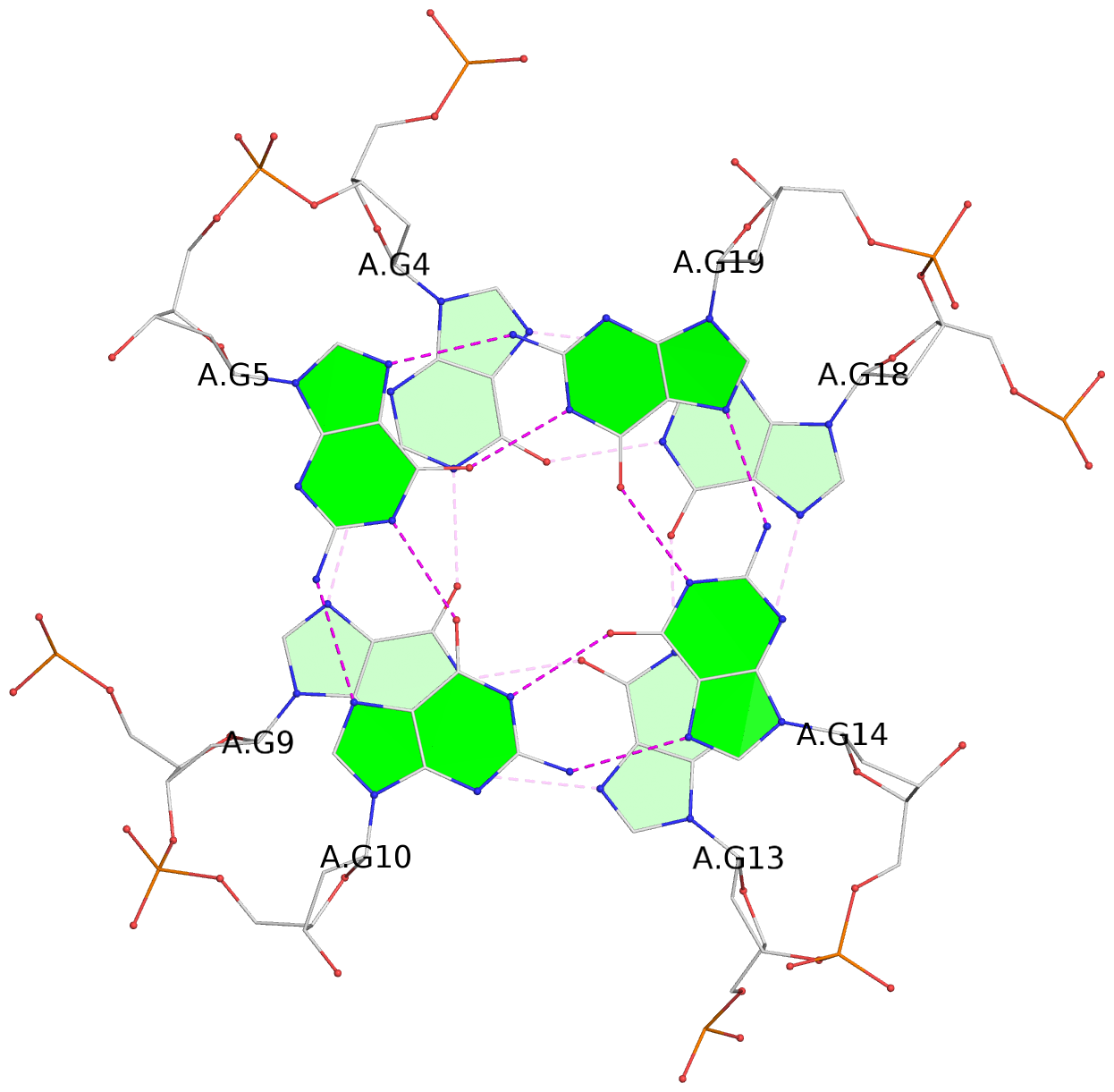
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.129 type=planar nts=4 GGGG A.DG3,A.DG8,A.DG12,A.DG17 2 glyco-bond=---- sugar=---- groove=---- planarity=0.207 type=other nts=4 GGGG A.DG4,A.DG9,A.DG13,A.DG18 3 glyco-bond=---- sugar=---- groove=---- planarity=0.228 type=other nts=4 GGGG A.DG5,A.DG10,A.DG14,A.DG19
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.