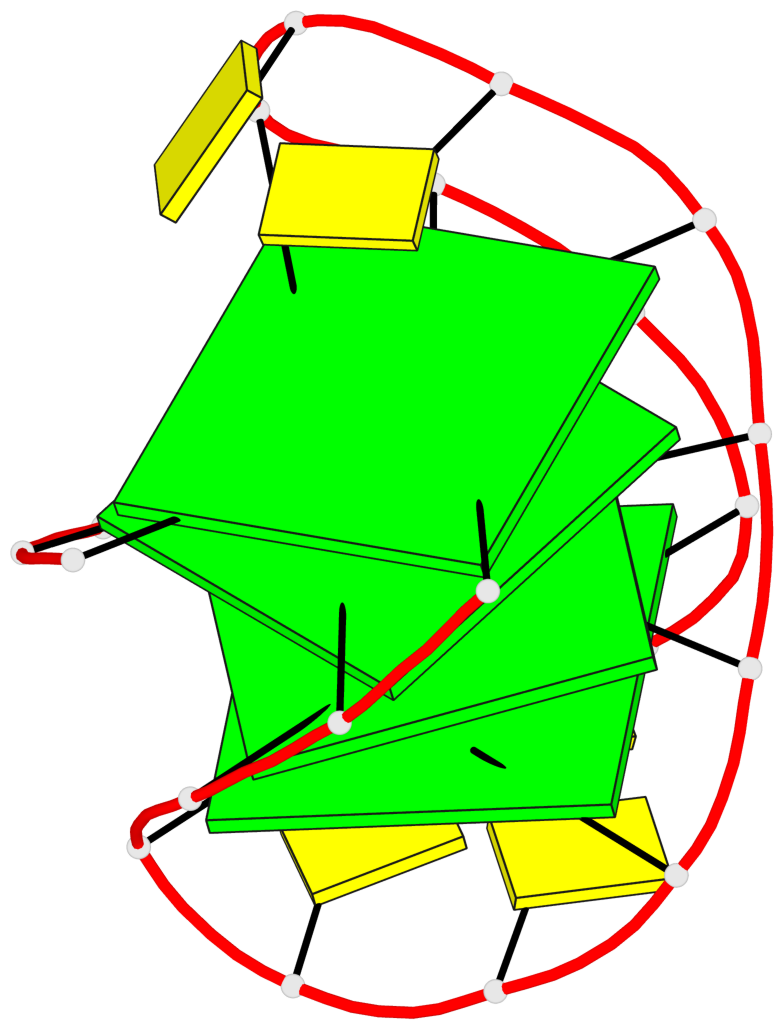
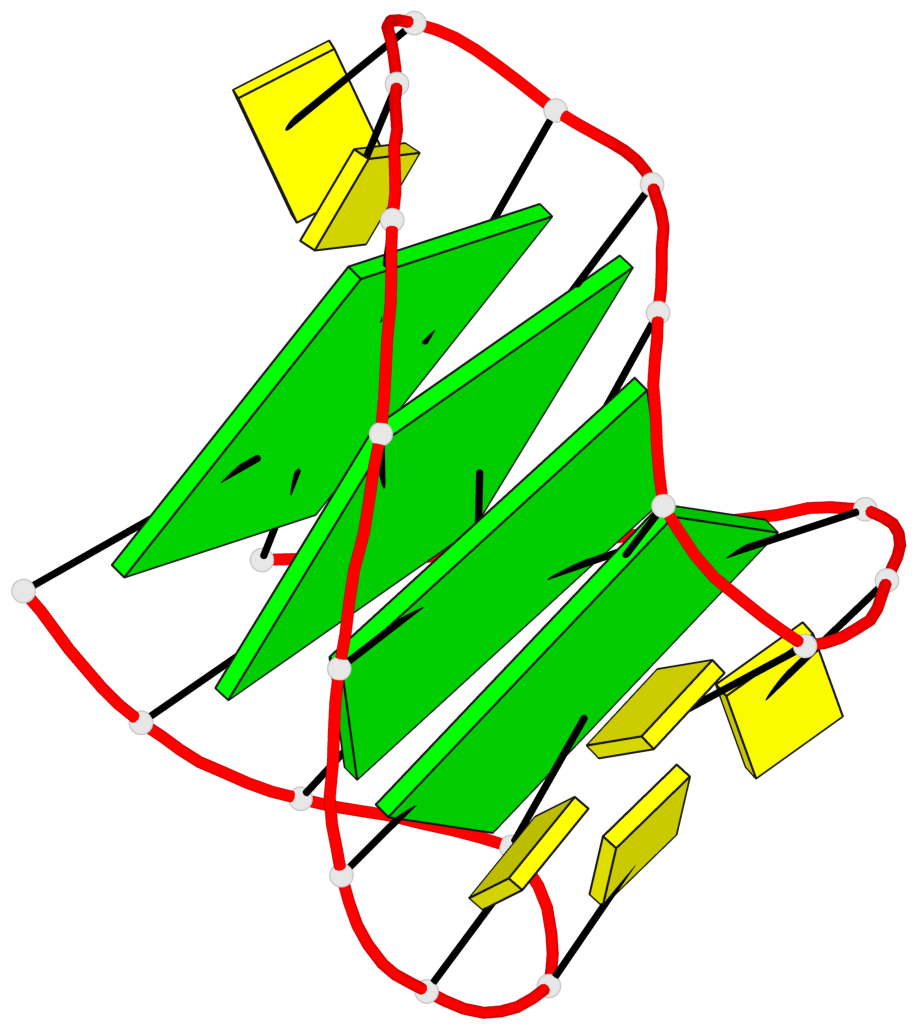
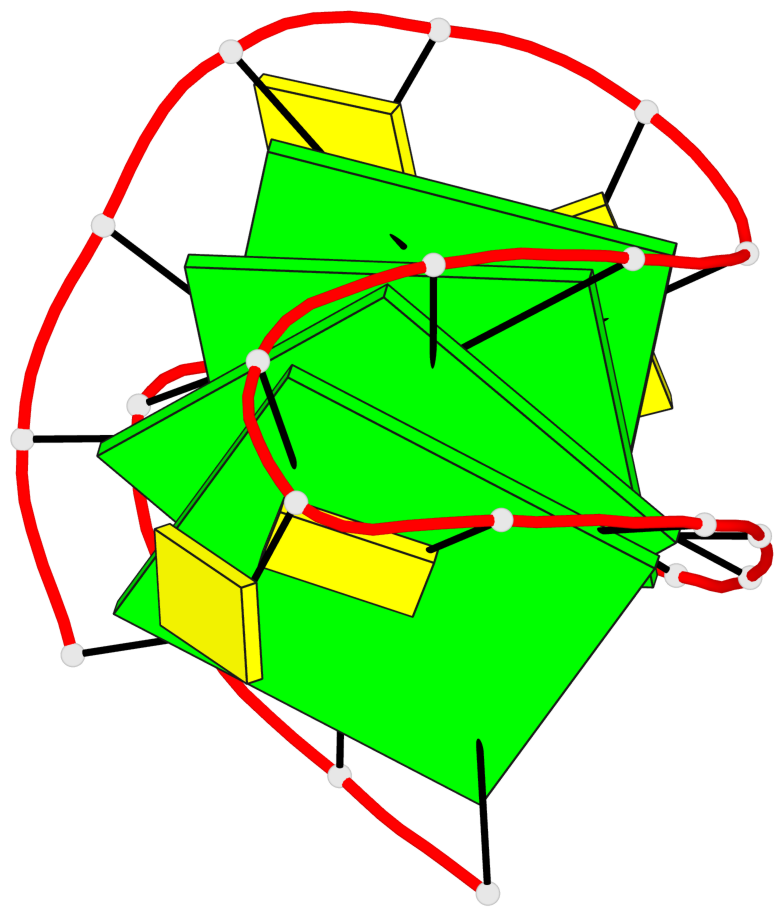
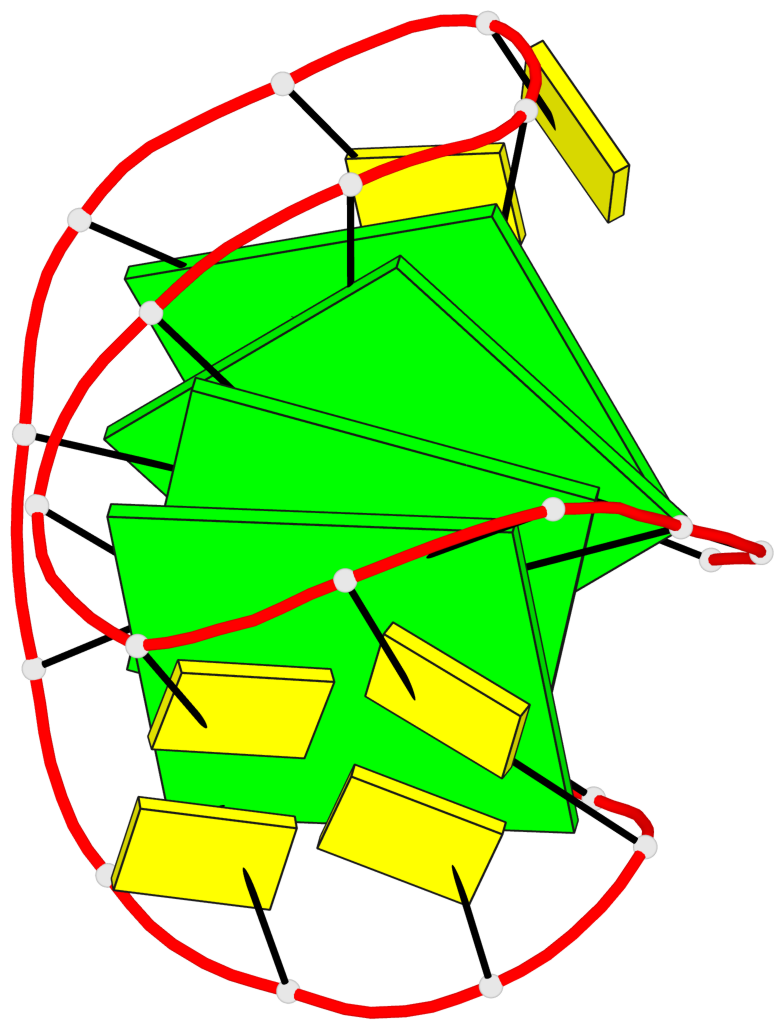
Detailed DSSR results for the G-quadruplex: PDB entry 5oph
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 5oph
- Class
- DNA
- Method
- NMR
- Summary
- G-quadruplex structure of DNA oligonucleotide containing ggggcc repeats linked to als and ftd
- Reference
- Brcic J, Plavec J (2018): "NMR structure of a G-quadruplex formed by four d(G4C2) repeats: insights into structural polymorphism." Nucleic Acids Res., 46, 11605-11617. doi: 10.1093/nar/gky886.
- Abstract
- Most frequent genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), is a largely increased number of d(G4C2)n•(G2C4)n repeats located in the non-coding region of C9orf72 gene. Non-canonical structures, including G-quadruplexes, formed within expanded repeats have been proposed to drive repeat expansion and pathogenesis of ALS and FTD. Oligonucleotide d[(G4C2)3G4], which represents the shortest oligonucleotide model of d(G4C2) repeats with the ability to form a unimolecular G-quadruplex, forms two major G-quadruplex structures in addition to several minor species which coexist in solution with K+ ions. Herein, we used solution-state NMR to determine the high-resolution structure of one of the major G-quadruplex species adopted by d[(G4C2)3G4]. Structural characterization of the G-quadruplex named AQU was facilitated by a single substitution of dG with 8Br-dG at position 21 and revealed an antiparallel fold composed of four G-quartets and three lateral C-C loops. The G-quadruplex exhibits high thermal stability and is favored kinetically and under slightly acidic conditions. An unusual structural element distinct from a C-quartet is observed in the structure. Two C•C base pairs are stacked on the nearby G-quartet and are involved in a dynamic equilibrium between symmetric N3-amino and carbonyl-amino geometries and protonated C+•C state.
- G4 notes
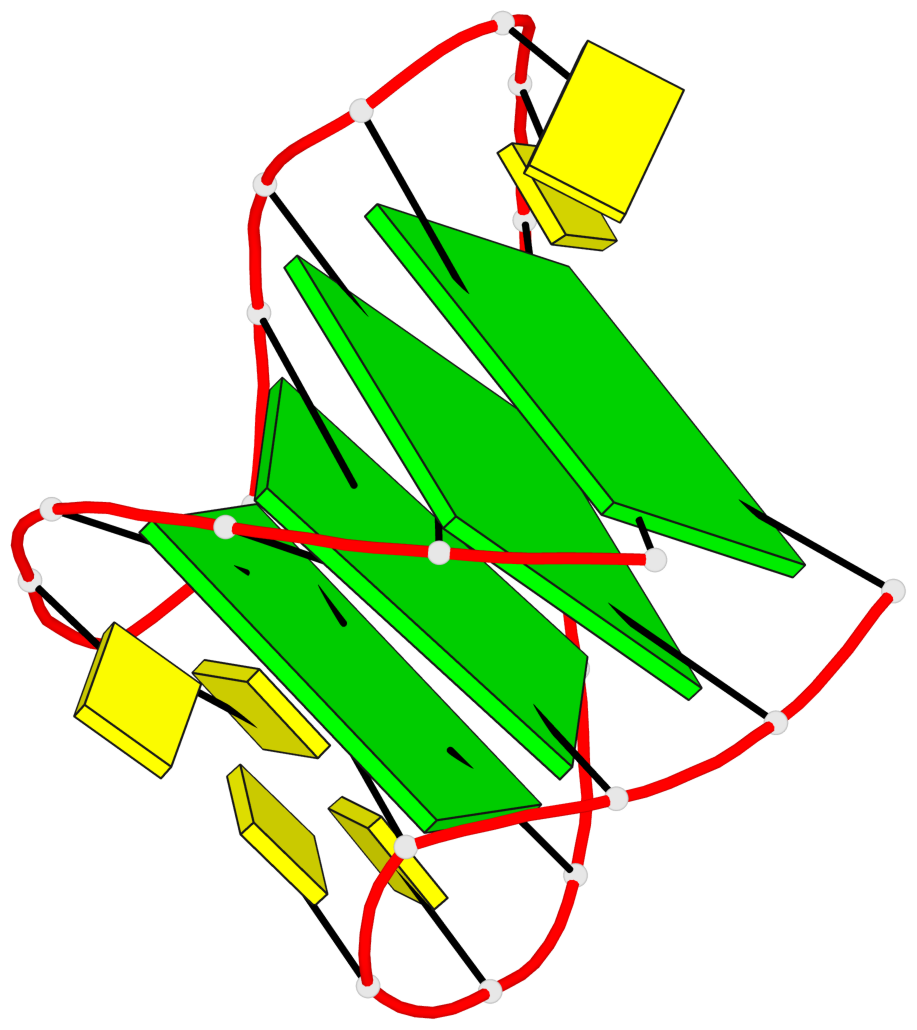
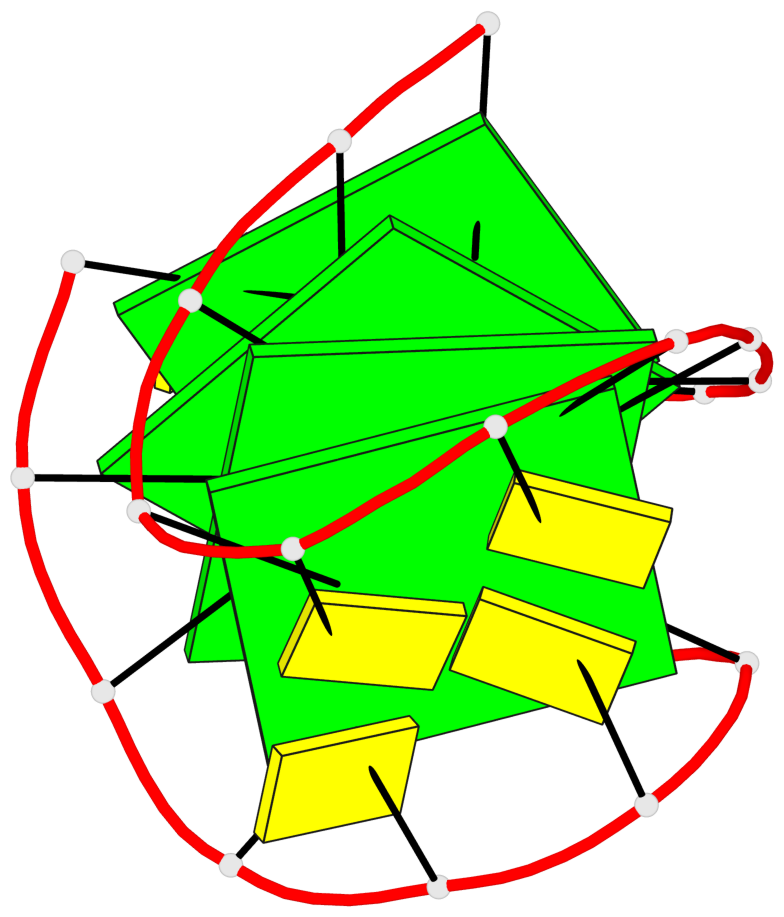
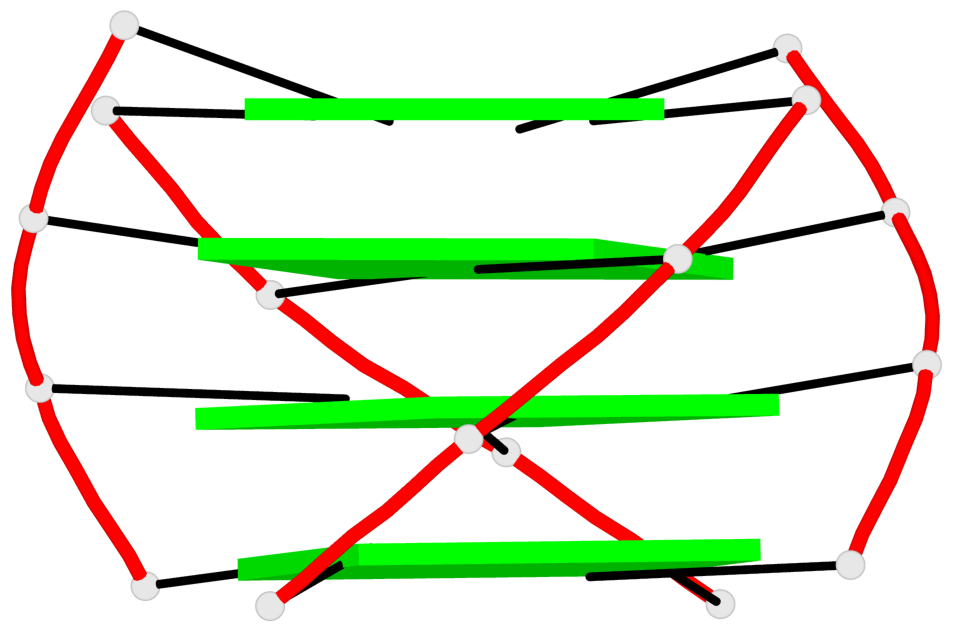
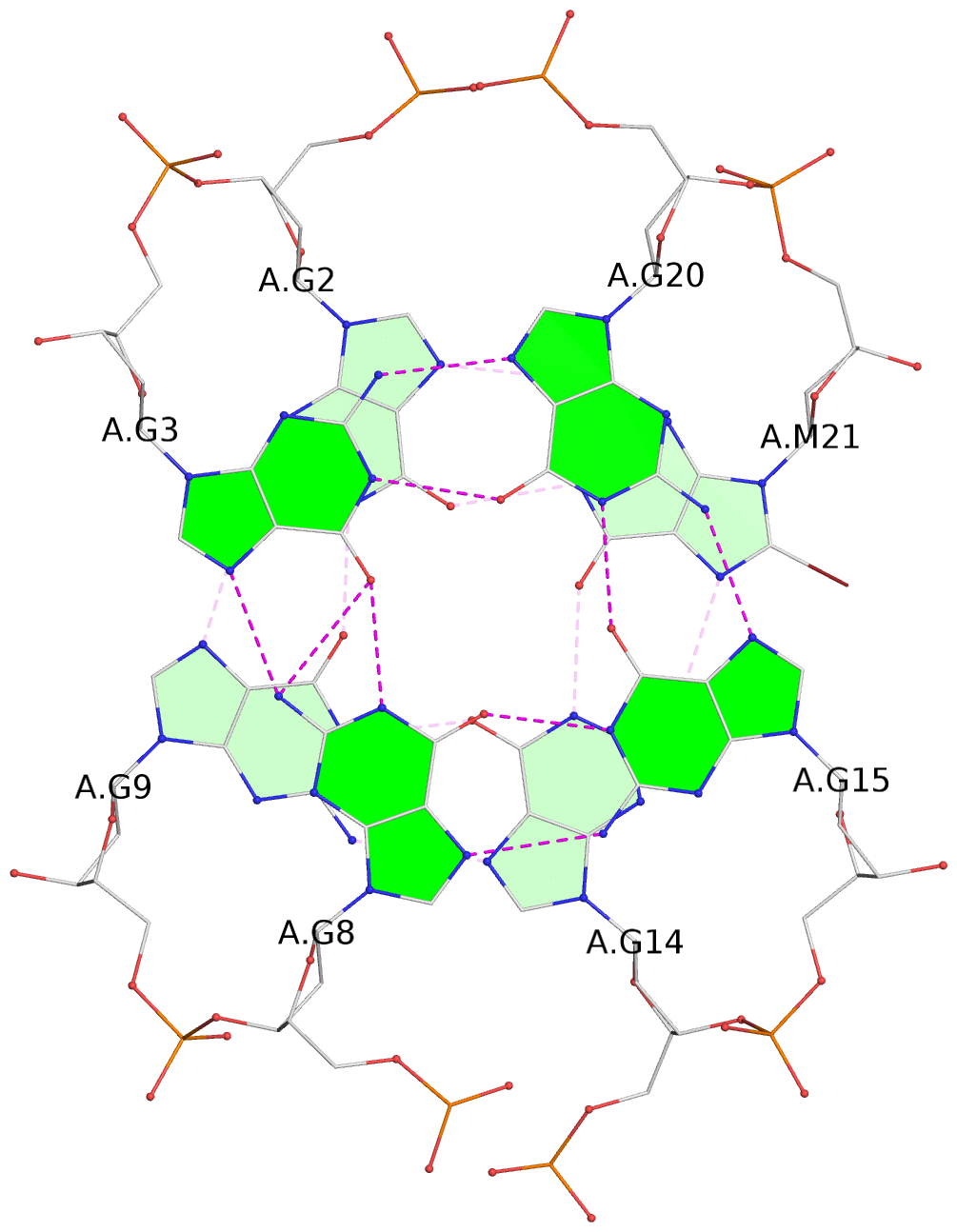
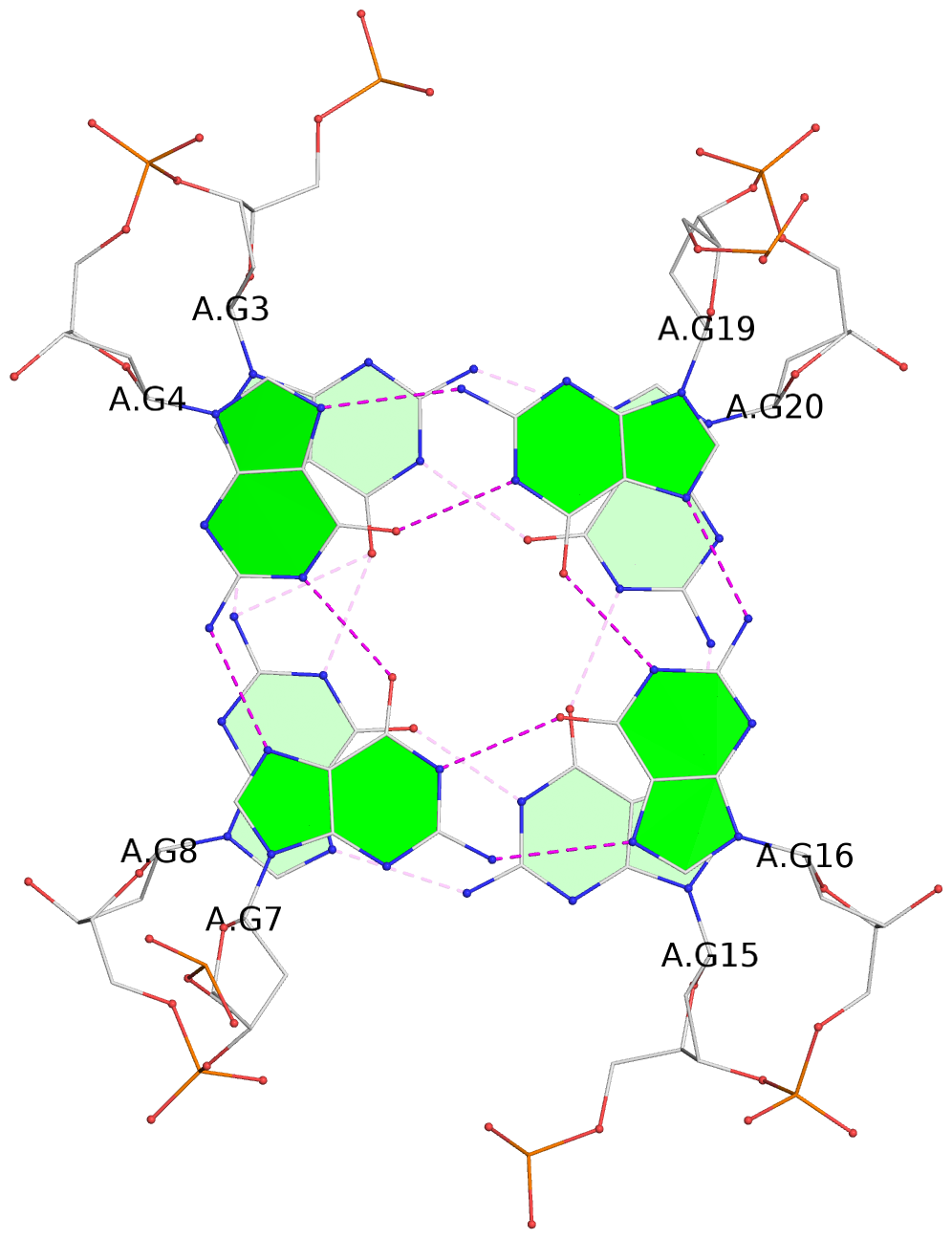
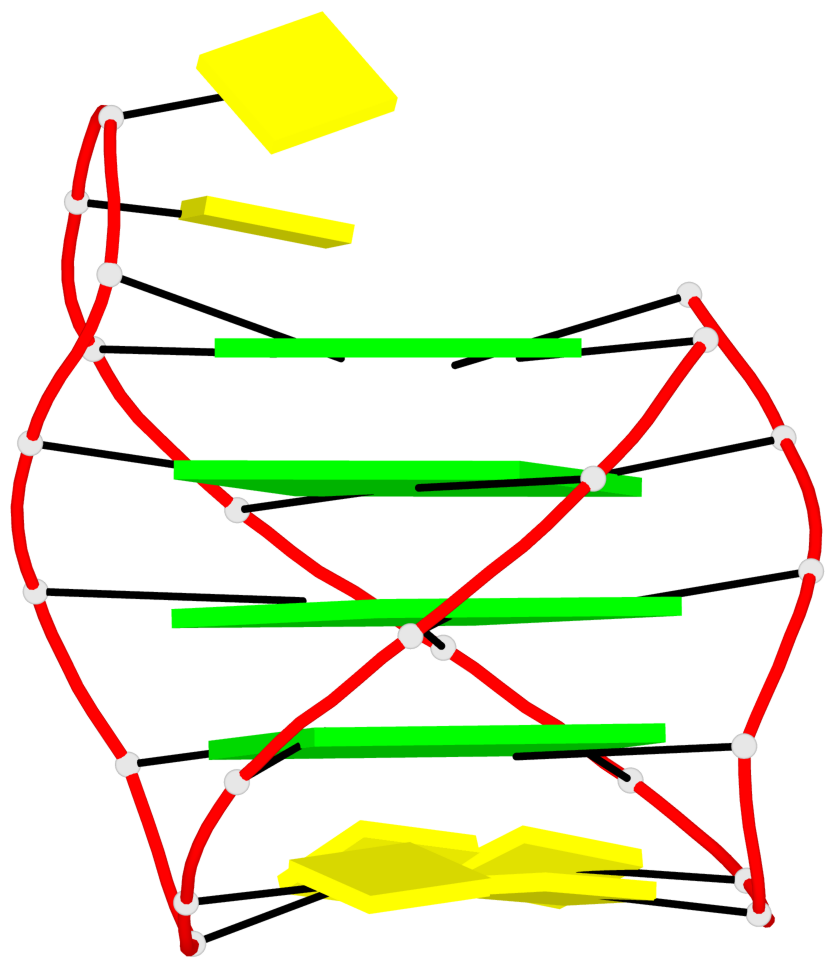
- 4 G-tetrads, 1 G4 helix, 1 G4 stem, 4(-Lw-Ln-Lw), chair(2+2), UDUD
Base-block schematics in six views
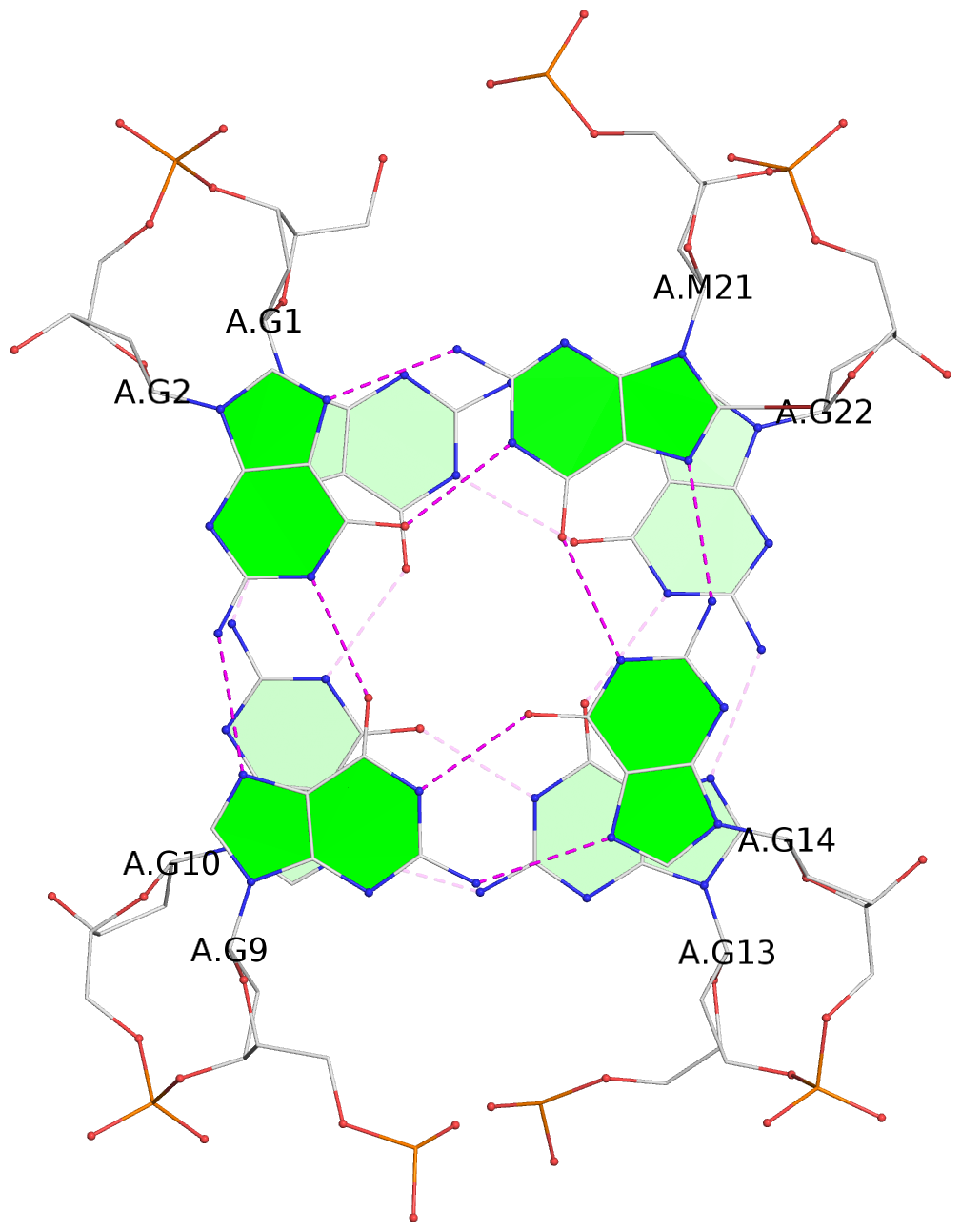
List of 4 G-tetrads
1 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.279 type=bowl nts=4 GGGG A.DG1,A.DG10,A.DG13,A.DG22 2 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.178 type=other nts=4 GGGg A.DG2,A.DG9,A.DG14,A.BGM21 3 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.176 type=other nts=4 GGGG A.DG3,A.DG8,A.DG15,A.DG20 4 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.292 type=other nts=4 GGGG A.DG4,A.DG7,A.DG16,A.DG19
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.