Detailed DSSR results for the G-quadruplex: PDB entry 5ov2
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 5ov2
- Class
- DNA
- Method
- NMR
- Summary
- 2'f-ana-g modified quadruplex with a flipped tetrad
- Reference
- Dickerhoff J, Weisz K (2017): "Nonconventional C-HF Hydrogen Bonds Support a Tetrad Flip in Modified G-Quadruplexes." J Phys Chem Lett, 8, 5148-5152. doi: 10.1021/acs.jpclett.7b02428.
- Abstract
- A G-quadruplex adopting a (3 + 1)-hybrid structure was substituted at its 5'-tetrad by 2'-deoxy-2'-fluoro-arabinoguanosine (FaraG) analogs. Incorporation of anti-favoring FaraG at syn-positions of the 5'-outer tetrad induced a reversal of the tetrad polarity without noticeably compromising the quadruplex stability. This conformational change is shown to be promoted by nonconventional C-H···F hydrogen bonds acting within the anti-FaraG residues.
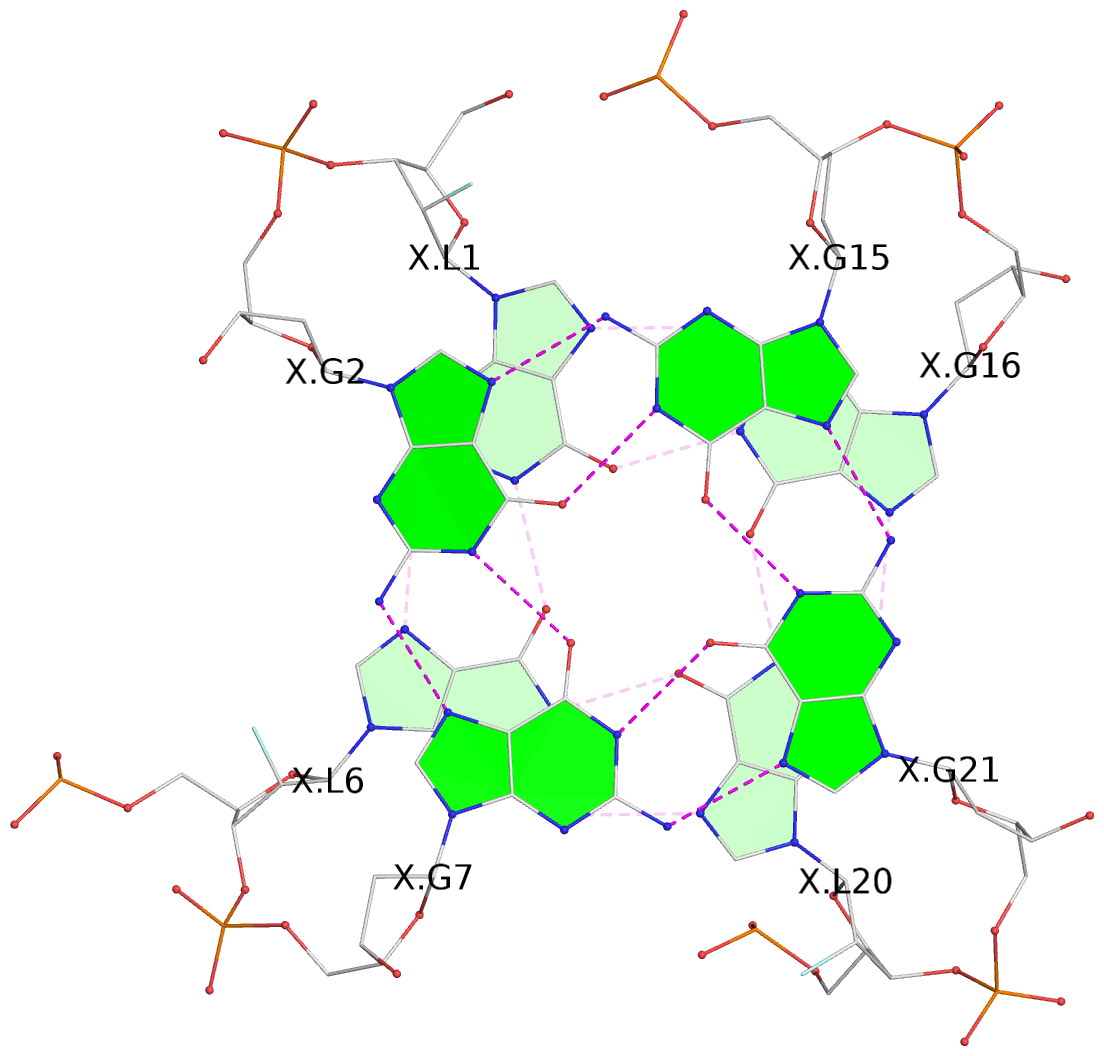
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-PD+Lw), U3D(3+1), UUUD
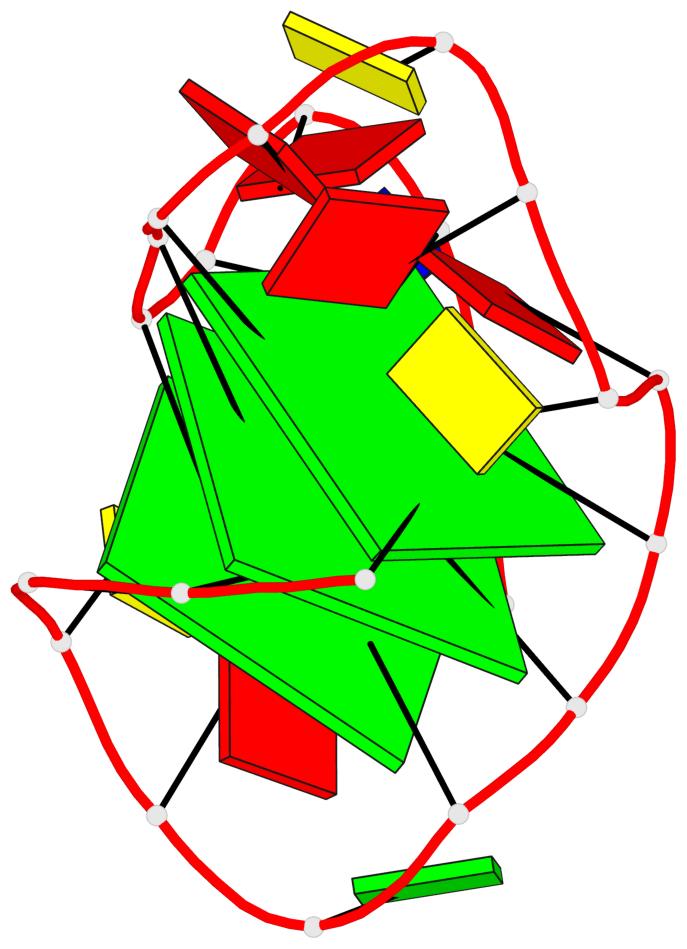
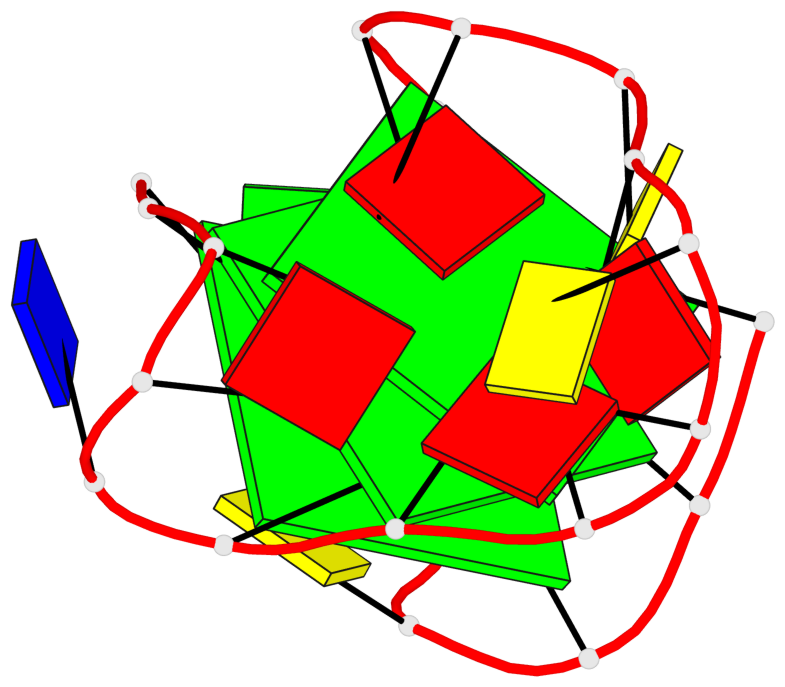
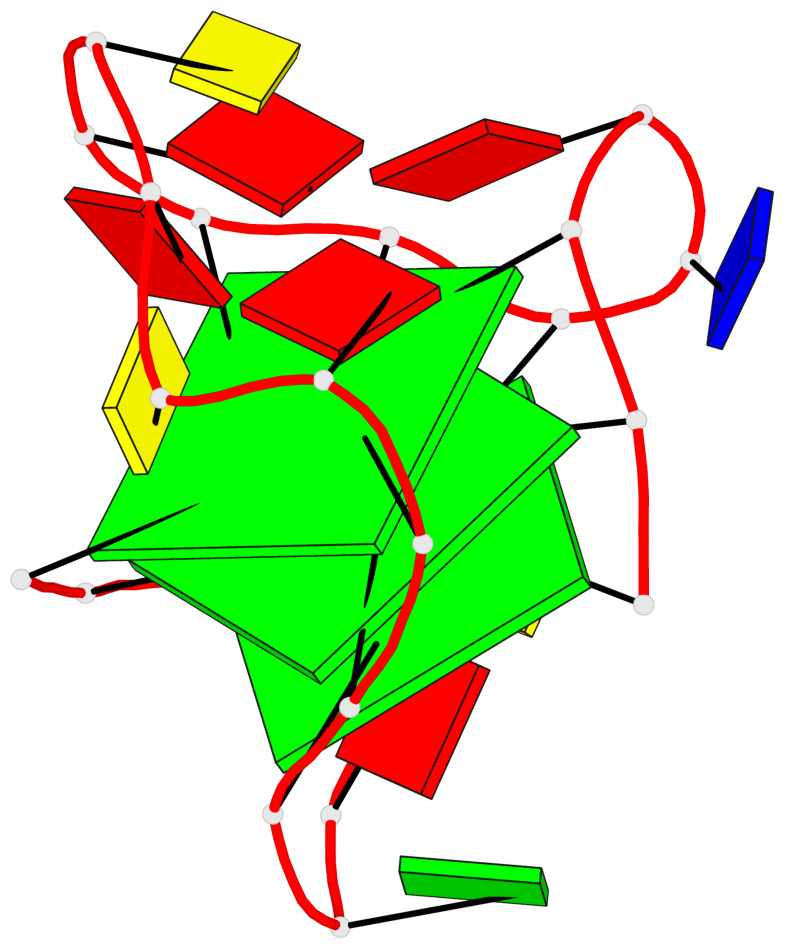
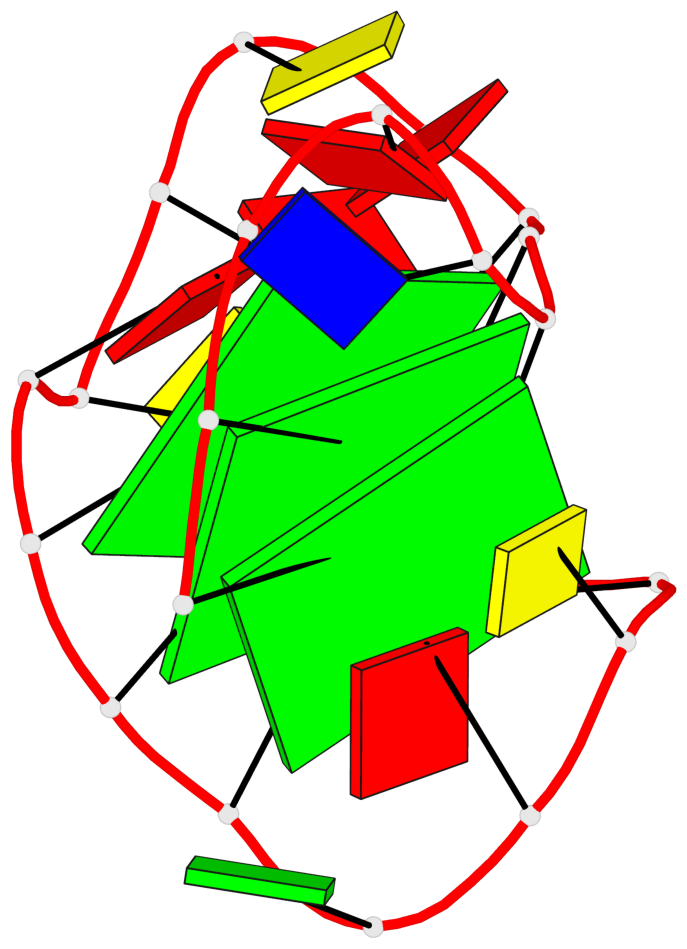
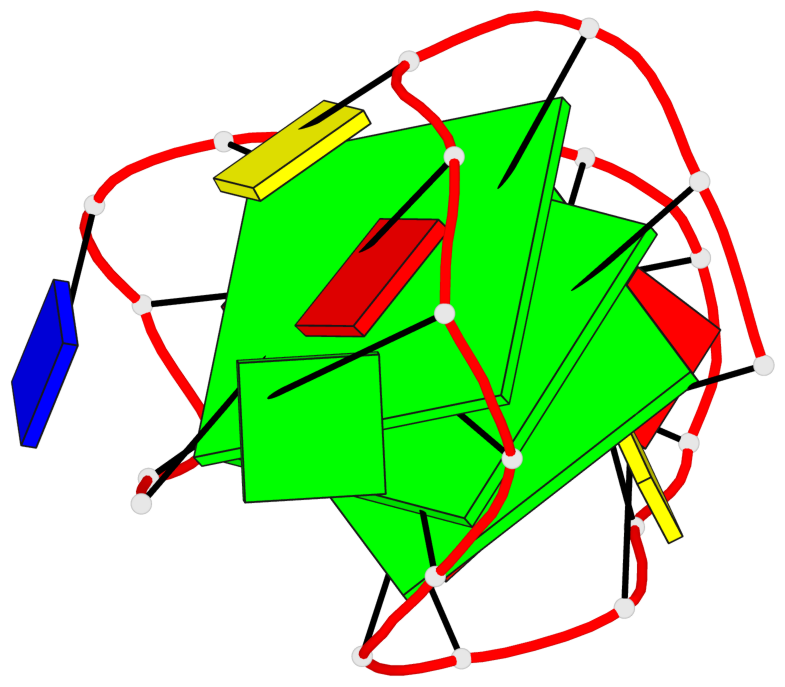
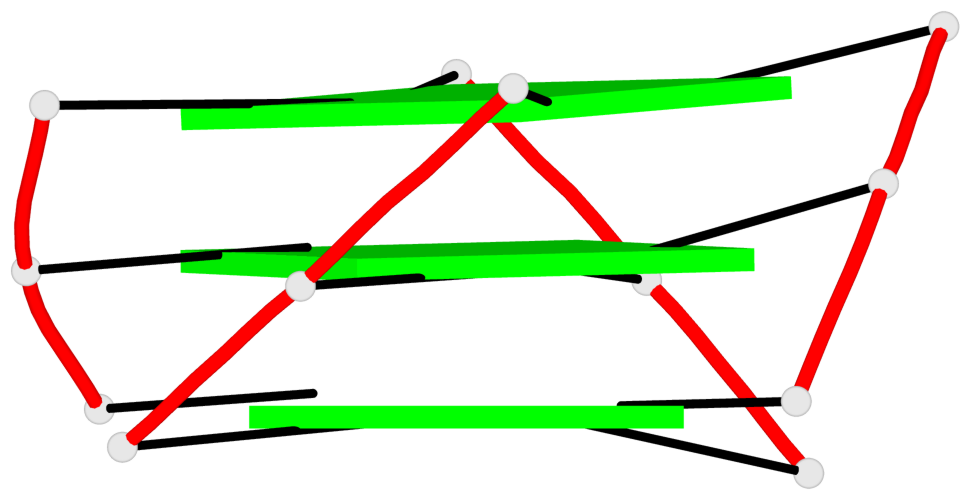
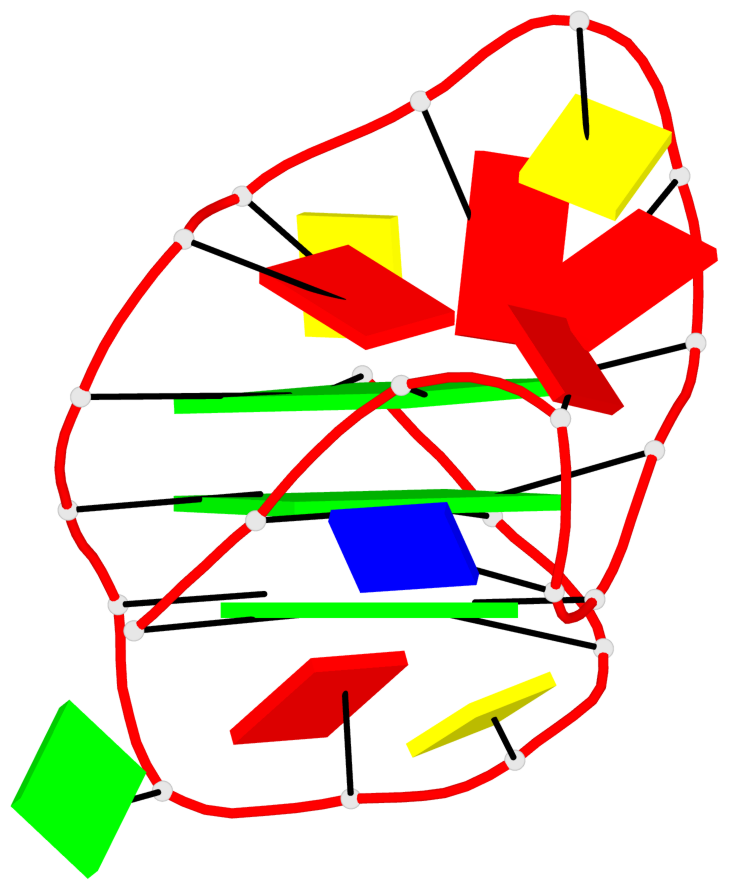
Base-block schematics in six views
List of 3 G-tetrads
1 glyco-bond=---s sugar=---- groove=--wn planarity=0.372 type=other nts=4 gggG X.GFL1,X.GFL6,X.GFL20,X.DG16 2 glyco-bond=---s sugar=---- groove=--wn planarity=0.393 type=other nts=4 GGGG X.DG2,X.DG7,X.DG21,X.DG15 3 glyco-bond=---s sugar=.--- groove=--wn planarity=0.279 type=saddle nts=4 GGGG X.DG3,X.DG8,X.DG22,X.DG14
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.