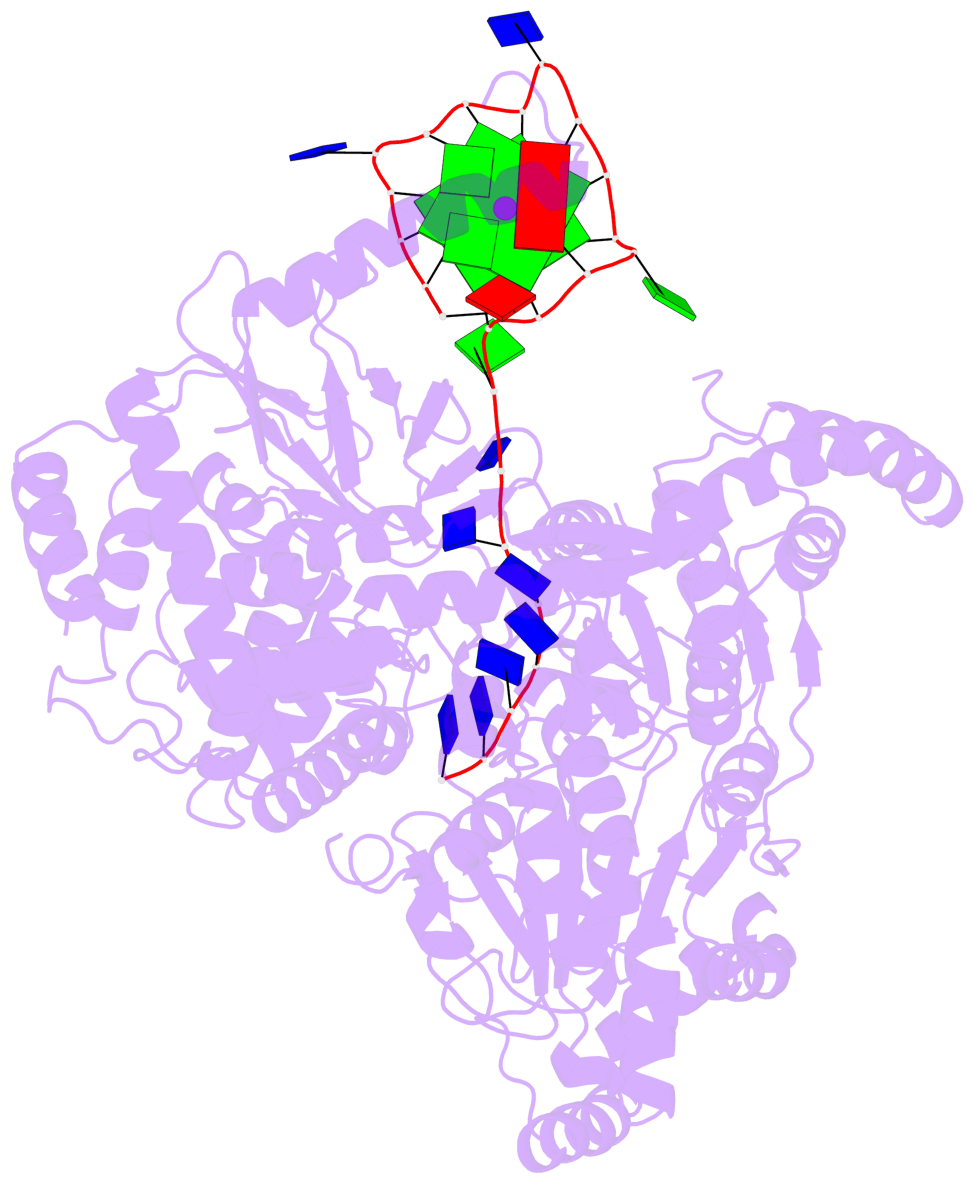
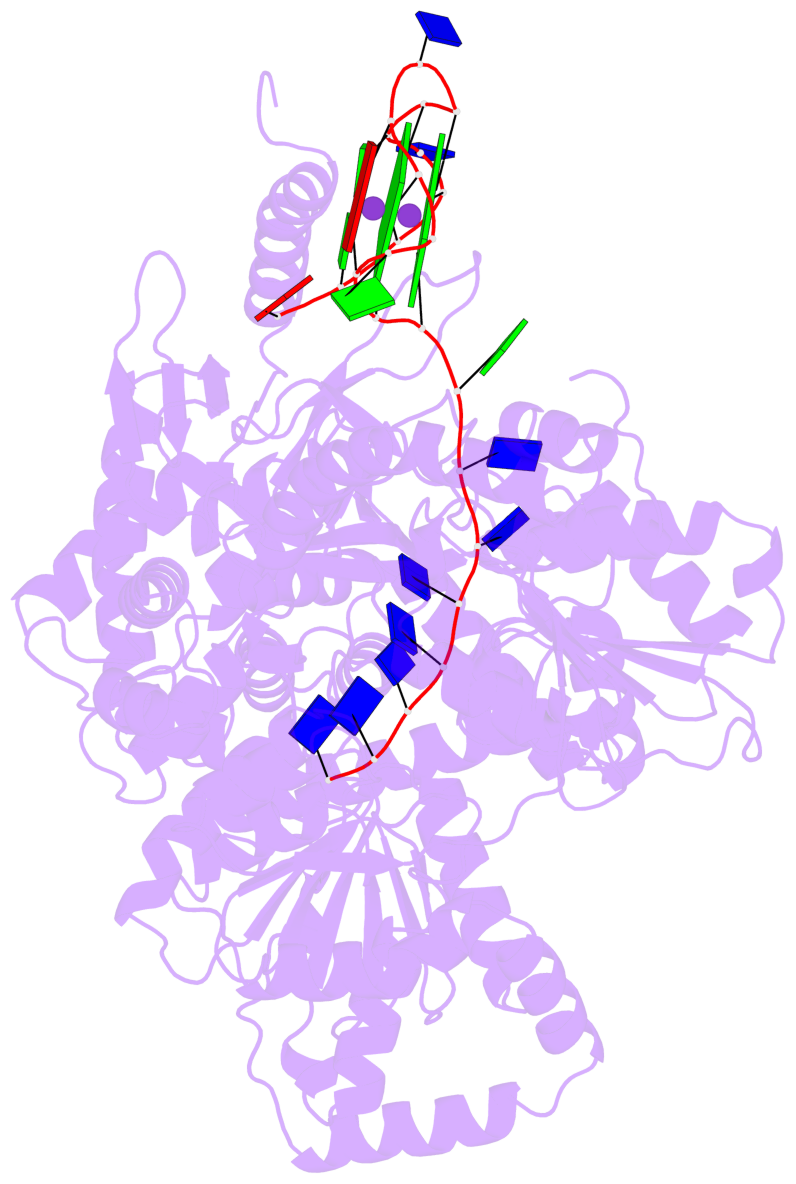
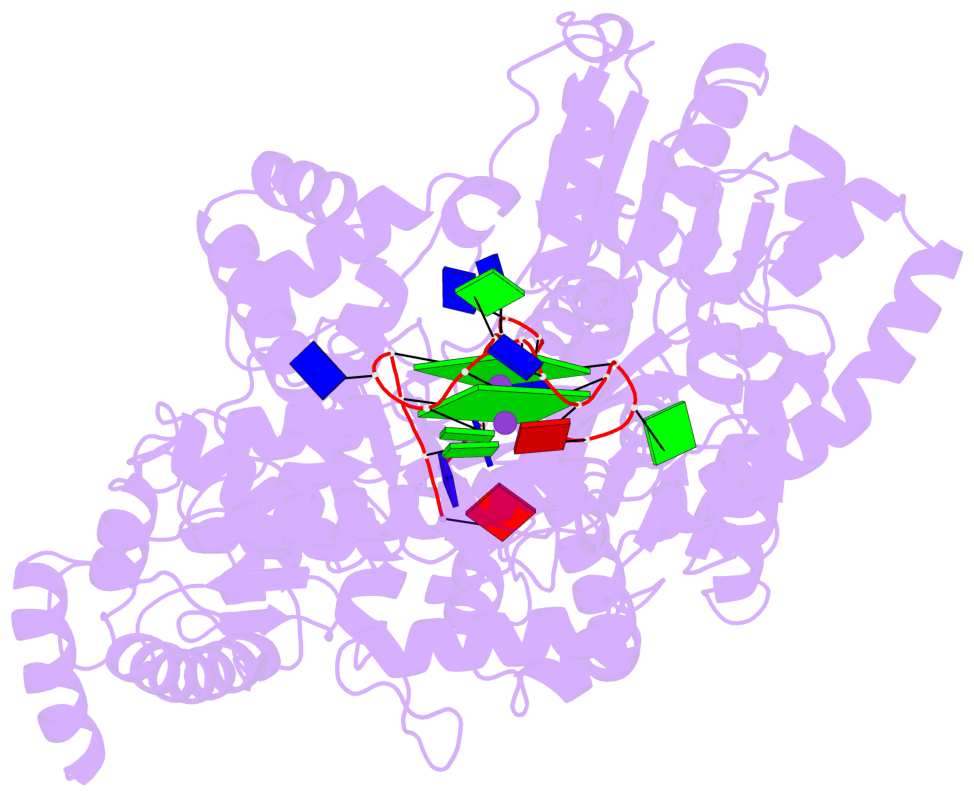
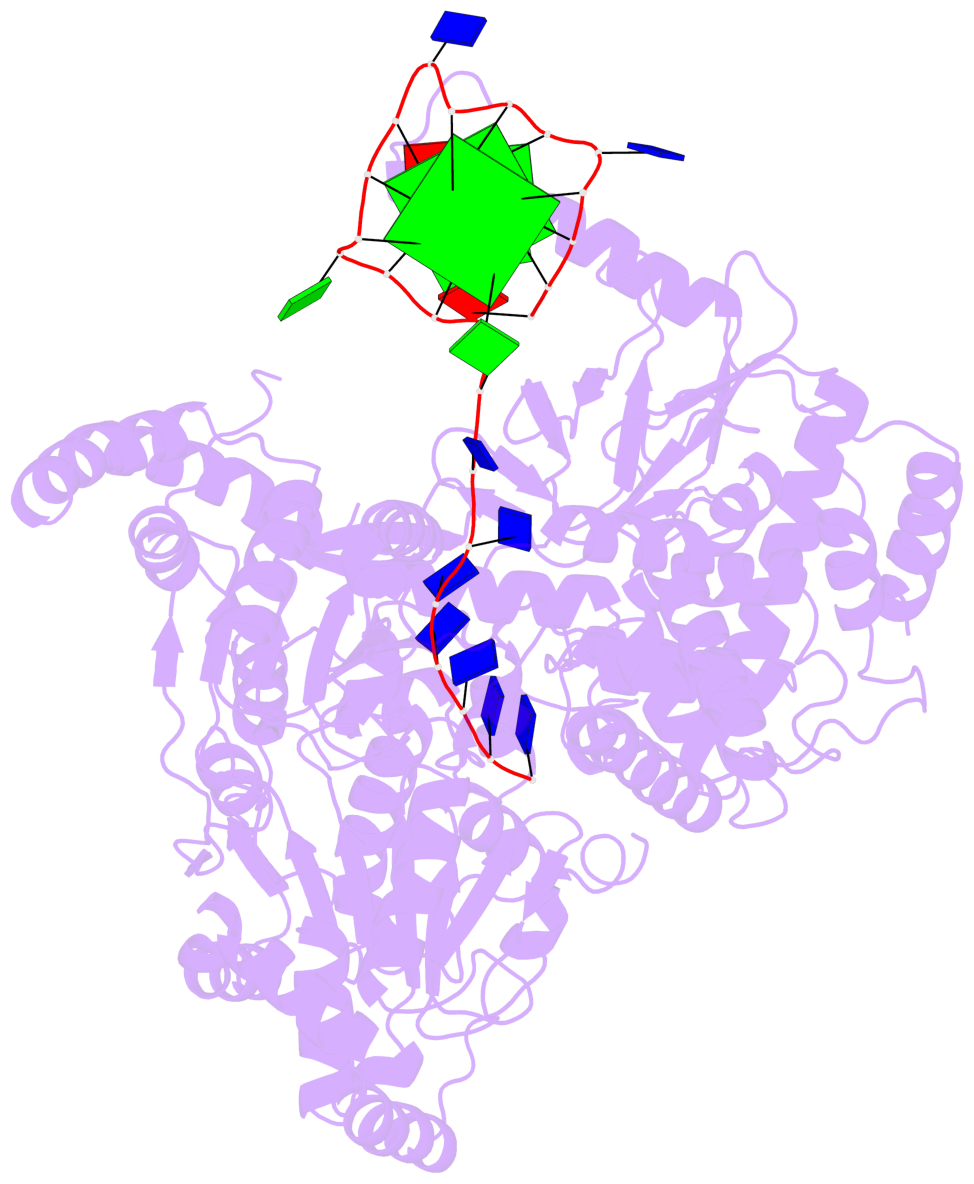
Detailed DSSR results for the G-quadruplex: PDB entry 5vhe
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 5vhe
- Class
- hydrolase
- Method
- X-ray (3.793 Å)
- Summary
- Dhx36 in complex with the c-myc g-quadruplex
- Reference
- Chen MC, Tippana R, Demeshkina NA, Murat P, Balasubramanian S, Myong S, Ferre-D'Amare AR (2018): "Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36." Nature, 558, 465-469. doi: 10.1038/s41586-018-0209-9.
- Abstract
- Guanine-rich nucleic acid sequences challenge the replication, transcription, and translation machinery by spontaneously folding into G-quadruplexes, the unfolding of which requires forces greater than most polymerases can exert1,2. Eukaryotic cells contain numerous helicases that can unfold G-quadruplexes 3 . The molecular basis of the recognition and unfolding of G-quadruplexes by helicases remains poorly understood. DHX36 (also known as RHAU and G4R1), a member of the DEAH/RHA family of helicases, binds both DNA and RNA G-quadruplexes with extremely high affinity4-6, is consistently found bound to G-quadruplexes in cells7,8, and is a major source of G-quadruplex unfolding activity in HeLa cell lysates 6 . DHX36 is a multi-functional helicase that has been implicated in G-quadruplex-mediated transcriptional and post-transcriptional regulation, and is essential for heart development, haematopoiesis, and embryogenesis in mice9-12. Here we report the co-crystal structure of bovine DHX36 bound to a DNA with a G-quadruplex and a 3' single-stranded DNA segment. We show that the N-terminal DHX36-specific motif folds into a DNA-binding-induced α-helix that, together with the OB-fold-like subdomain, selectively binds parallel G-quadruplexes. Comparison with unliganded and ATP-analogue-bound DHX36 structures, together with single-molecule fluorescence resonance energy transfer (FRET) analysis, suggests that G-quadruplex binding alone induces rearrangements of the helicase core; by pulling on the single-stranded DNA tail, these rearrangements drive G-quadruplex unfolding one residue at a time.
- G4 notes
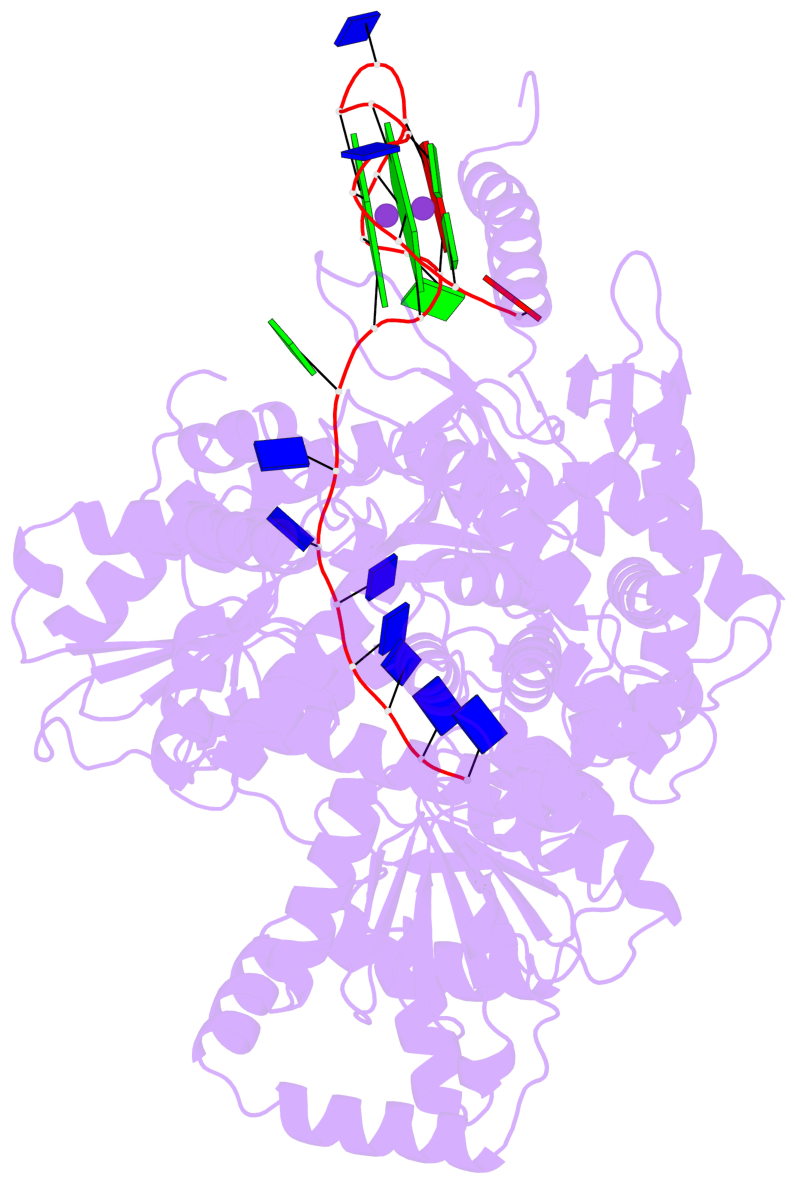
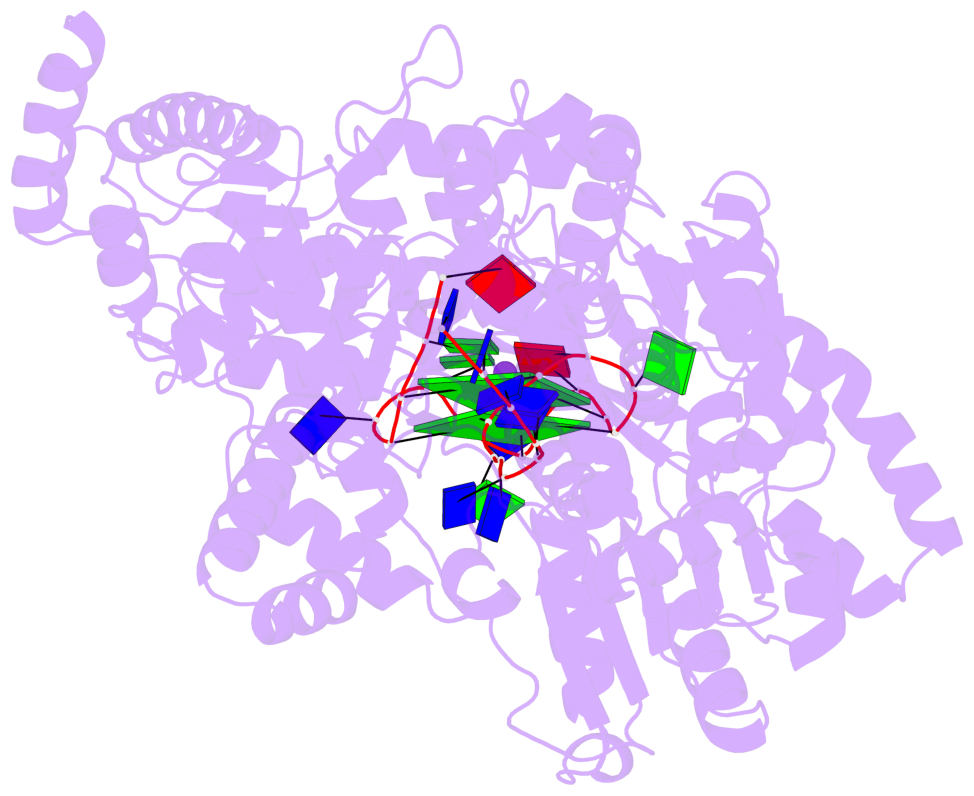
- 2 G-tetrads, 1 G4 helix, 1 G4 stem, 2(-P-P-P), parallel(4+0), UUUU
Base-block schematics in six views
List of 2 G-tetrads
1 glyco-bond=---- sugar=3-.- groove=---- planarity=0.219 type=other nts=4 GGGG B.DG3,B.DG7,B.DG11,B.DG15 2 glyco-bond=---- sugar=---3 groove=---- planarity=0.223 type=other nts=4 GGGG B.DG4,B.DG8,B.DG12,B.DG16
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.