Detailed DSSR results for the G-quadruplex: PDB entry 5z8f
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 5z8f
- Class
- DNA
- Method
- NMR
- Summary
- Solution structure for the unique dimeric 4:2 complex of a platinum(ii)-based tripod bound to a hybrid-1 human telomeric g-quadruplex
- Reference
- Liu W, Zhong YF, Liu LY, Shen CT, Zeng W, Wang F, Yang D, Mao ZW (2018): "Solution structures of multiple G-quadruplex complexes induced by a platinum(II)-based tripod reveal dynamic binding." Nat Commun, 9, 3496. doi: 10.1038/s41467-018-05810-4.
- Abstract
- DNA G-quadruplexes are not only attractive drug targets for cancer therapeutics, but also have important applications in supramolecular assembly. Here, we report a platinum(II)-based tripod (Pt-tripod) specifically binds the biological relevant hybrid-1 human telomeric G-quadruplex (Tel26), and strongly inhibits telomerase activity. Further investigations illustrate Pt-tripod induces the formation of monomeric and multimeric Pt-tripod‒Tel26 complex structures in solution. We solve the 1:1 and the unique dimeric 4:2 Pt-tripod-Tel26 complex structures by NMR. The structures indicate preferential binding of Pt-tripod to the 5'-end of Tel26 at a low Pt-tripod/Tel26 ratio of 0-1.0. After adding more Pt-tripod, the Pt-tripod binds the 3'-end of Tel26, unexpectedly inducing a unique dimeric 4:2 structure interlocked by an A:A non-canonical pair at the 3'-end. Our structures provide a structural basis for understanding the dynamic binding of small molecules with G-quadruplex and DNA damage mechanisms, and insights into the recognition and assembly of higher-order G-quadruplexes.
- G4 notes
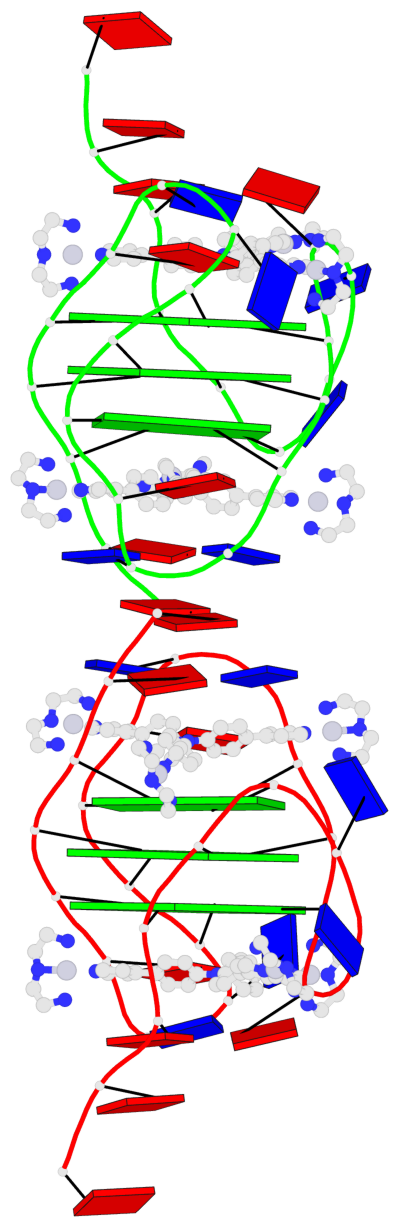
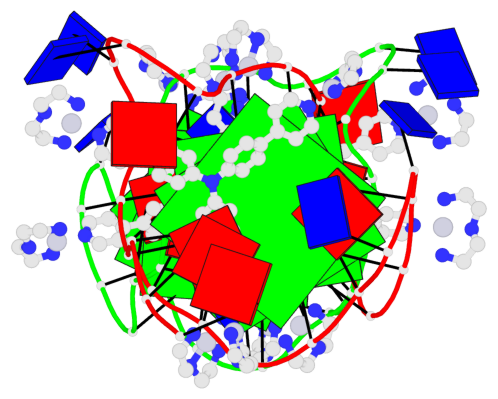
- 6 G-tetrads, 2 G4 helices, 2 G4 stems, 3(-P-Lw-Ln), hybrid-1(3+1), UUDU
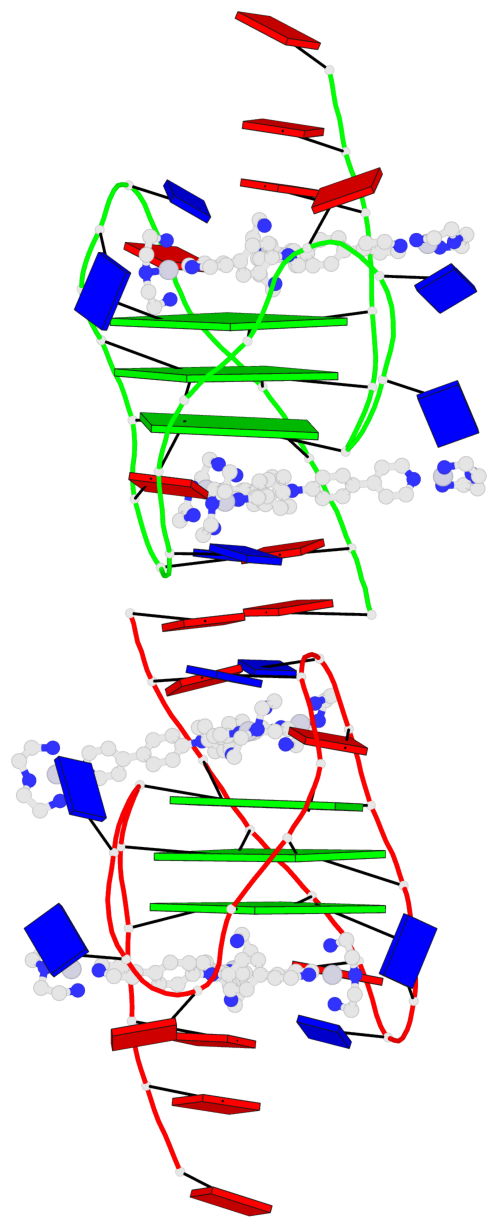
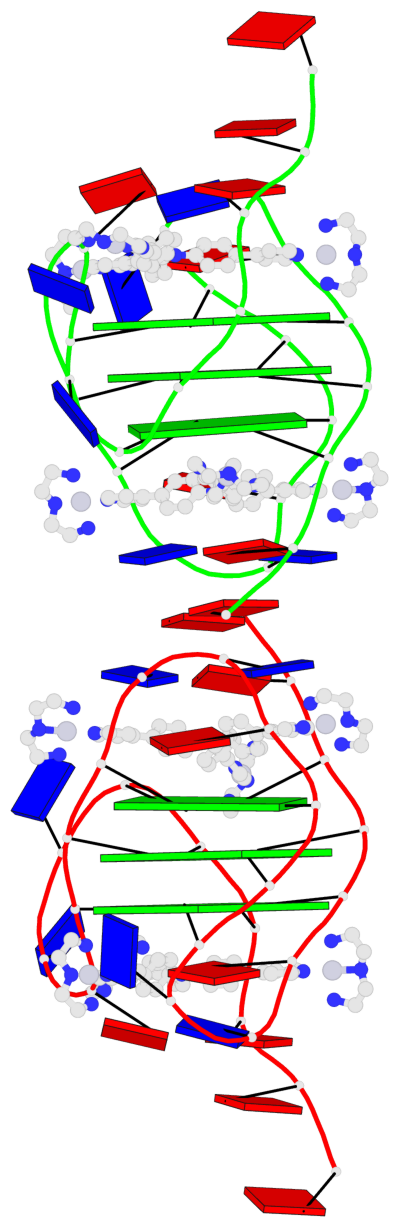
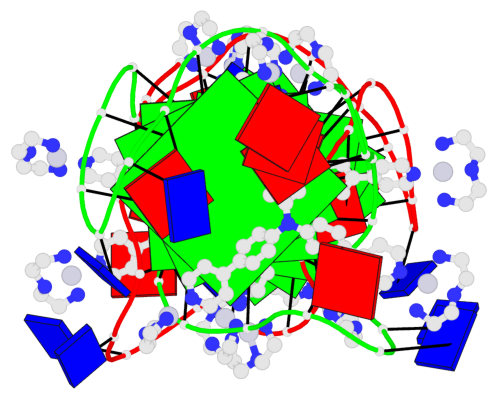
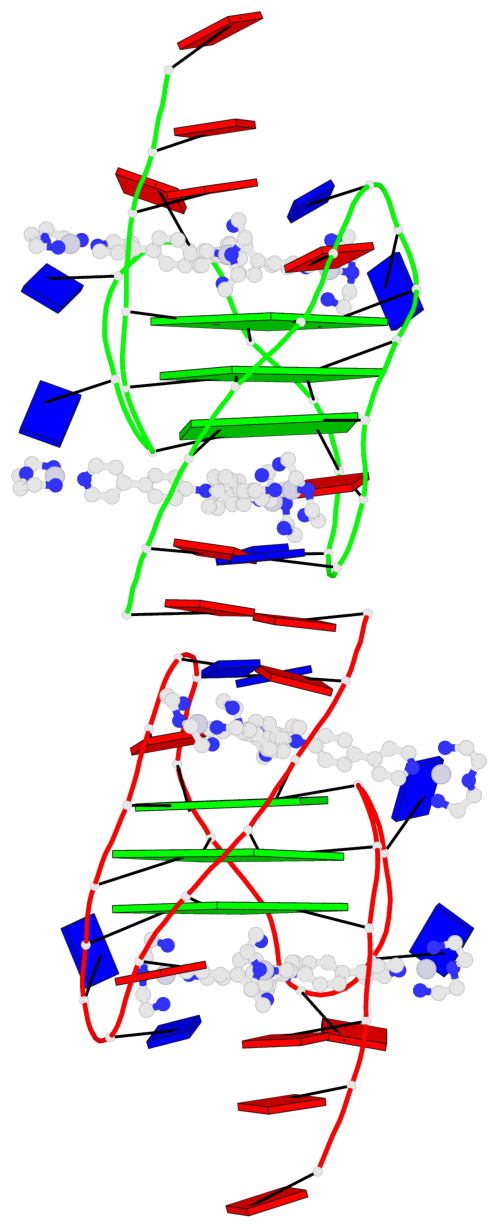
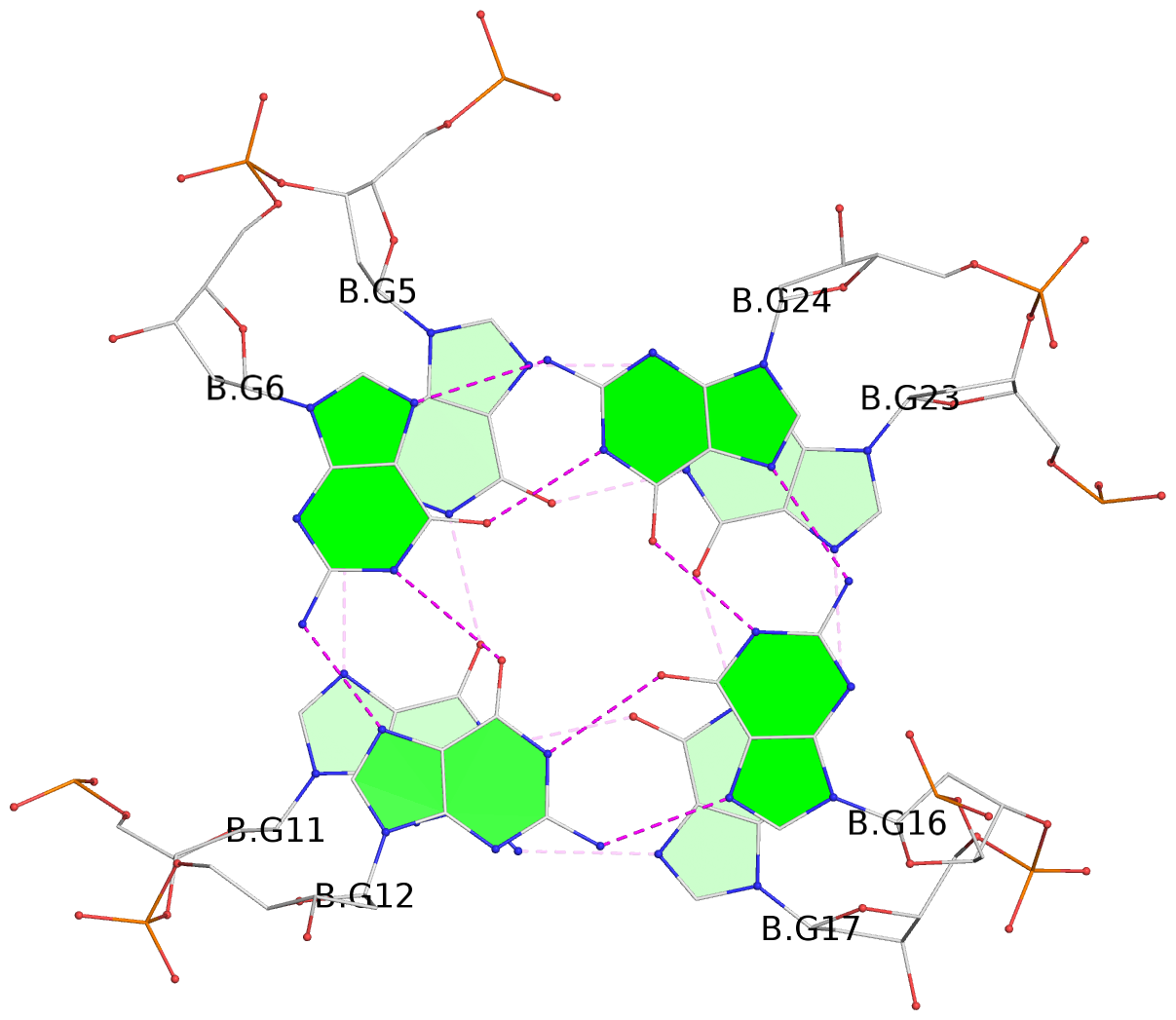
Base-block schematics in six views
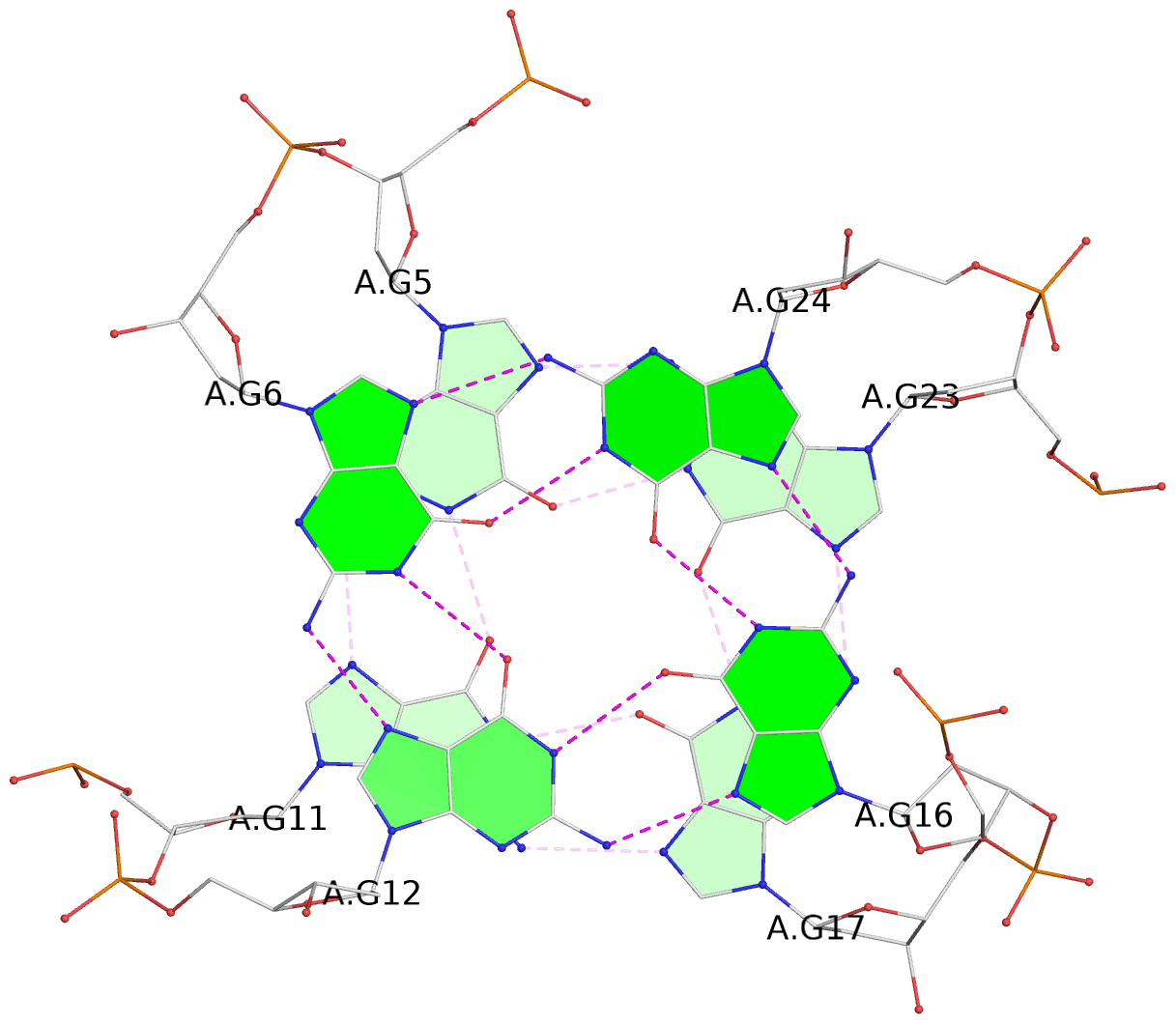
List of 6 G-tetrads
1 glyco-bond=ss-s sugar=--.- groove=-wn- planarity=0.407 type=saddle nts=4 GGGG A.DG4,A.DG10,A.DG18,A.DG22 2 glyco-bond=--s- sugar=..-. groove=-wn- planarity=0.348 type=other nts=4 GGGG A.DG5,A.DG11,A.DG17,A.DG23 3 glyco-bond=--s- sugar=-3-3 groove=-wn- planarity=0.297 type=bowl-2 nts=4 GGGG A.DG6,A.DG12,A.DG16,A.DG24 4 glyco-bond=ss-s sugar=--.- groove=-wn- planarity=0.354 type=saddle nts=4 GGGG B.DG4,B.DG10,B.DG18,B.DG22 5 glyco-bond=--s- sugar=..-. groove=-wn- planarity=0.272 type=other nts=4 GGGG B.DG5,B.DG11,B.DG17,B.DG23 6 glyco-bond=--s- sugar=-3-. groove=-wn- planarity=0.214 type=bowl-2 nts=4 GGGG B.DG6,B.DG12,B.DG16,B.DG24
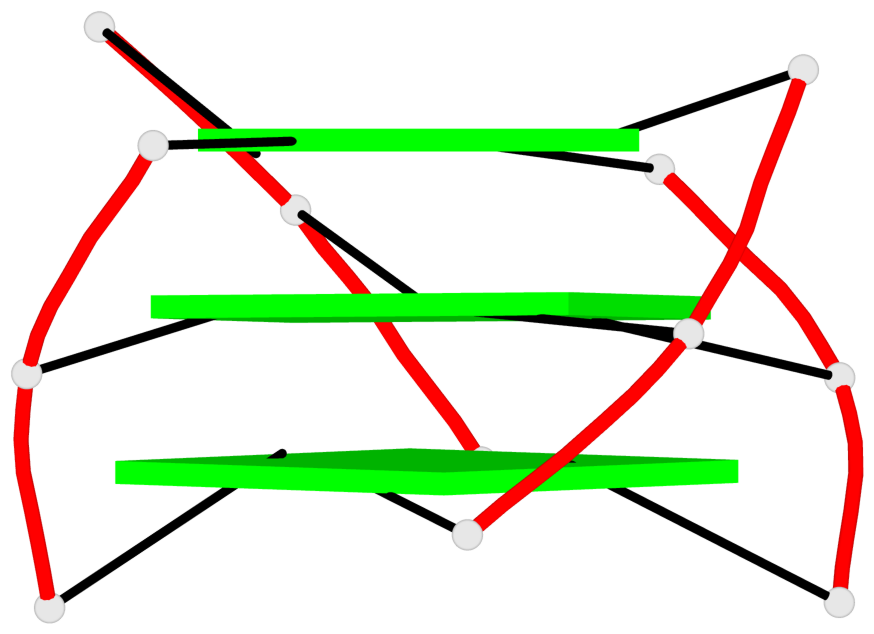
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
Helix#2, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.