Detailed DSSR results for the G-quadruplex: PDB entry 5zev
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 5zev
- Class
- DNA
- Method
- NMR
- Summary
- Solution structure of g-quadruplex formed in vegfr-2 proximal promoter sequence
- Reference
- Liu Y, Lan W, Wang C, Cao C (2018): "A putative G-quadruplex structure in the proximal promoter ofVEGFR-2has implications for drug design to inhibit tumor angiogenesis." J. Biol. Chem., 293, 8947-8955. doi: 10.1074/jbc.RA118.002666.
- Abstract
- Tumor angiogenesis is mainly regulated by vascular endothelial growth factor (VEGF) produced by cancer cells. It is active on the endothelium via VEGF receptor 2 (VEGFR-2). G-quadruplexes are DNA secondary structures formed by guanine-rich sequences, for example, within gene promoters where they may contribute to transcriptional activity. The proximal promoter of VEGFR-2 contains a G-quadruplex, which has been suggested to interact with small molecules that inhibit VEGFR-2 expression and thereby tumor angiogenesis. However, its structure is not known. Here, we determined its NMR solution structure, which is composed of three stacked G-tetrads containing three syn guanines. The first guanine (G1) is positioned within the central G-tetrad. We also observed that a noncanonical, V-shaped loop spans three G-tetrad planes, including no bridging nucleotides. A long and diagonal loop, which includes six nucleotides, connects reversal double chains. With a melting temperature of 54.51 °C, the scaffold of this quadruplex is stabilized by one G-tetrad plane stacking with one nonstandard bp, G3-C8, whose bases interact with each other through only one hydrogen bond. In summary, the NMR solution structure of the G-quadruplex in the proximal promoter region of the VEGFR-2 gene reported here has uncovered its key features as a potential anticancer drug target.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 2(D+PX), UD3(1+3), UDDD
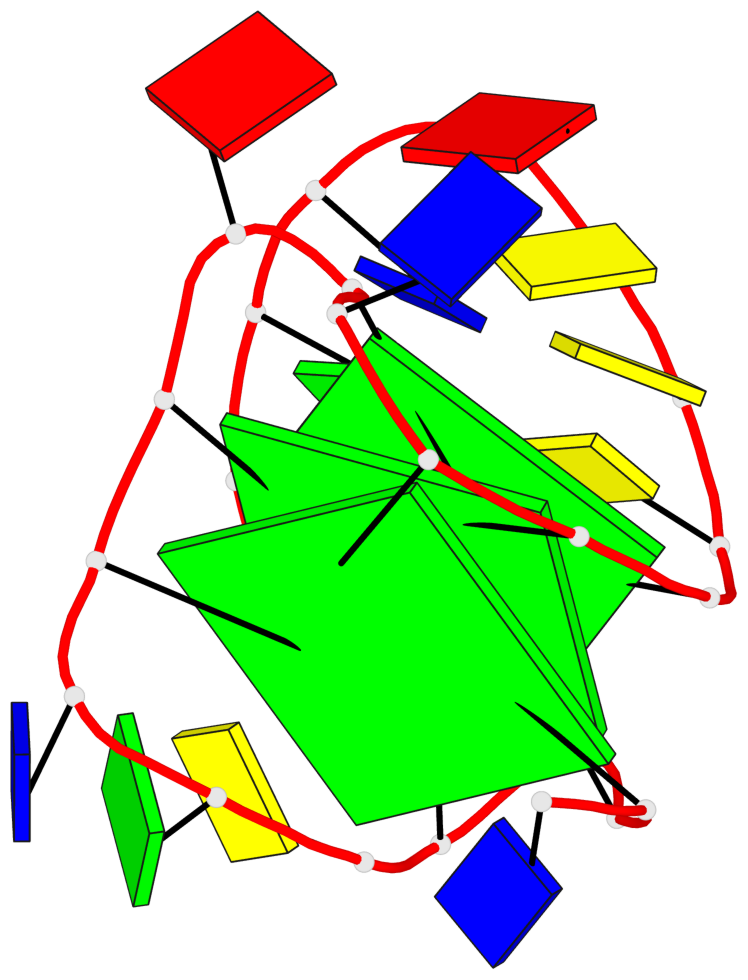
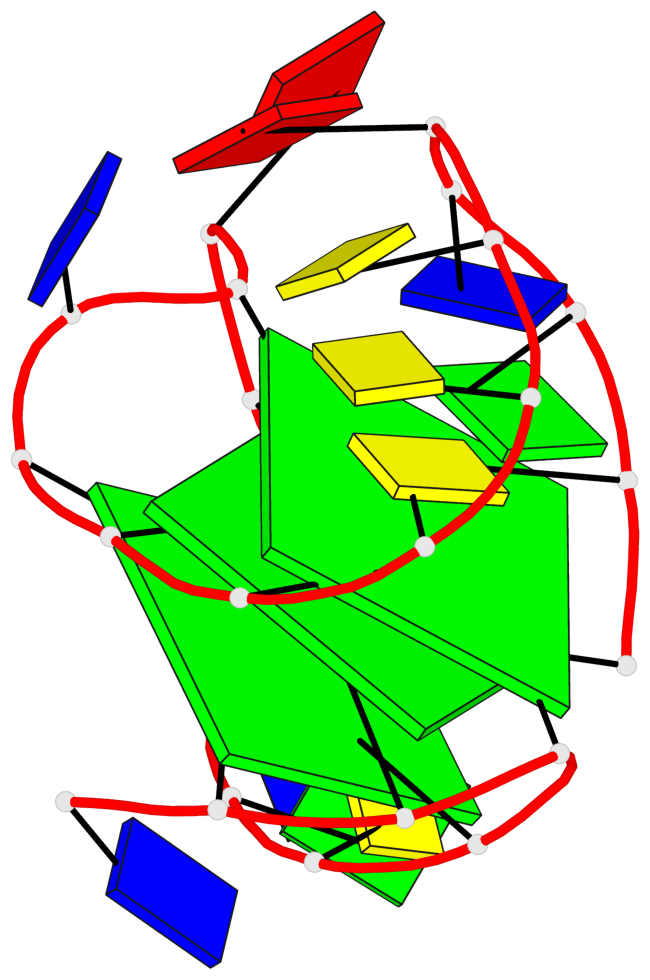
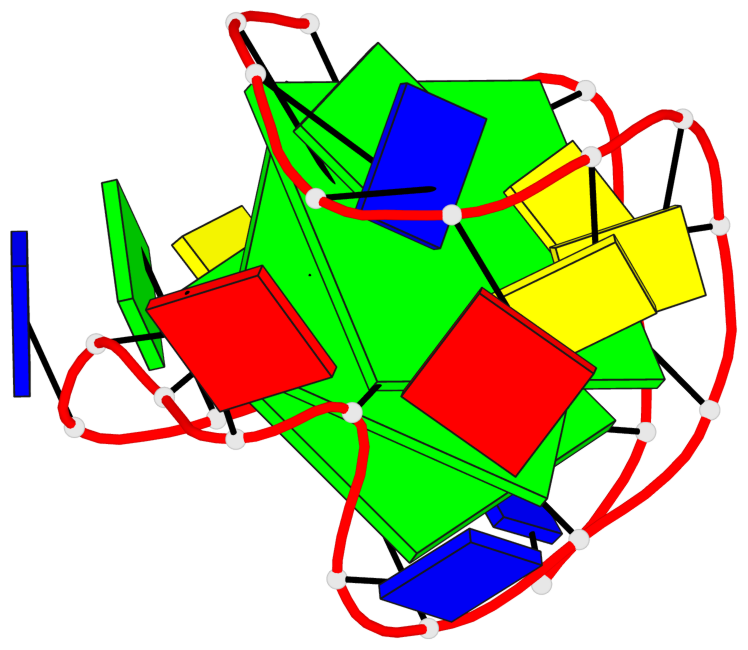
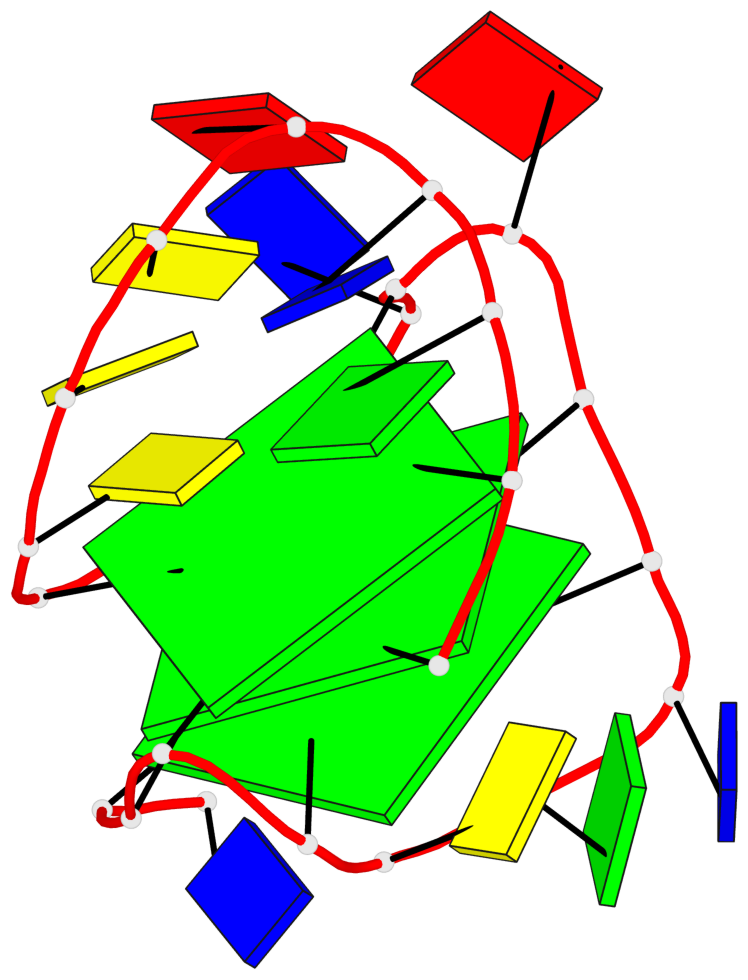
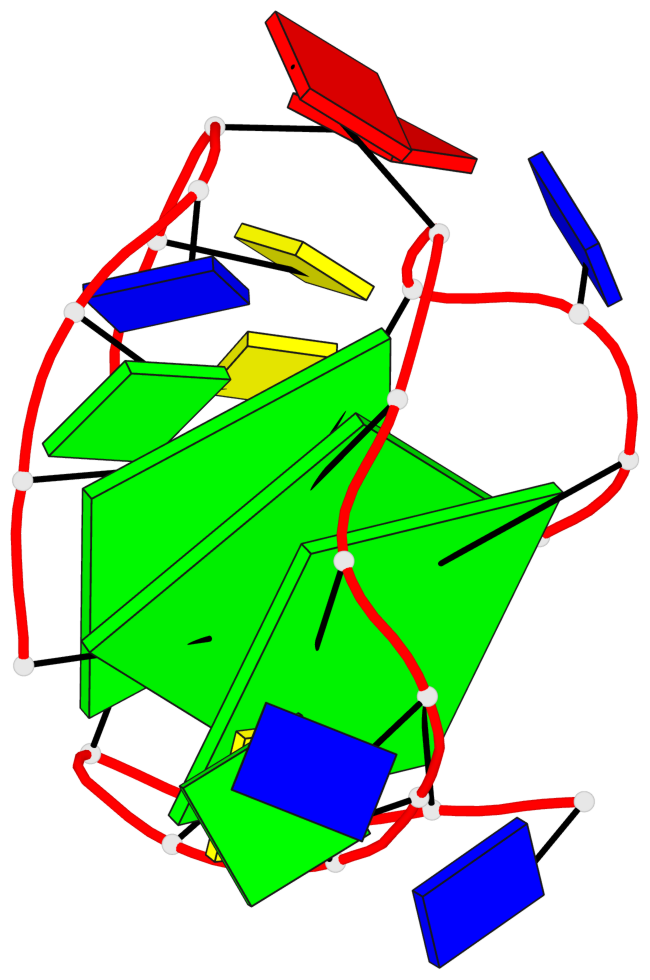
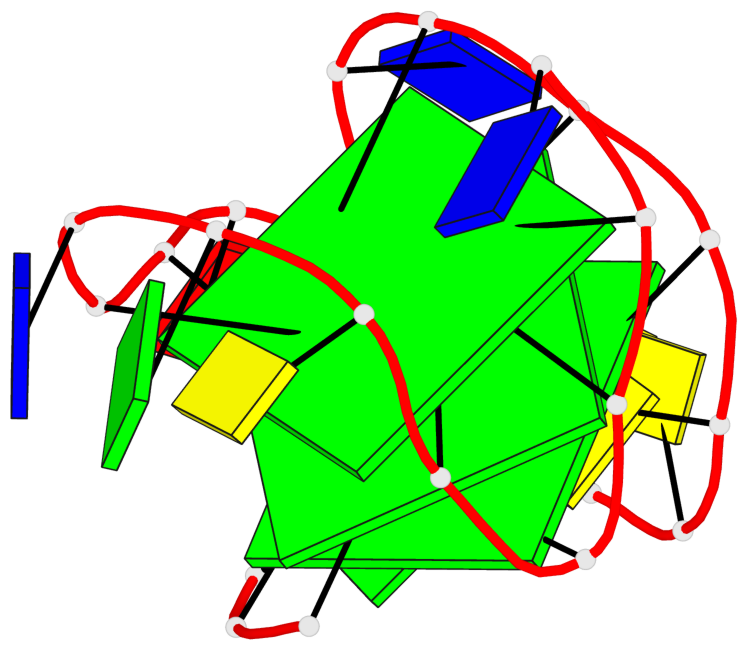
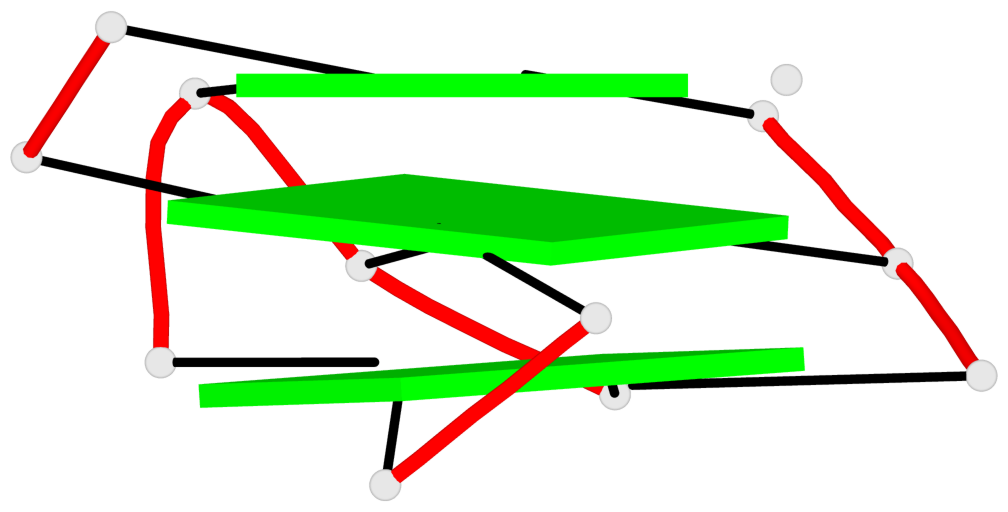
Base-block schematics in six views
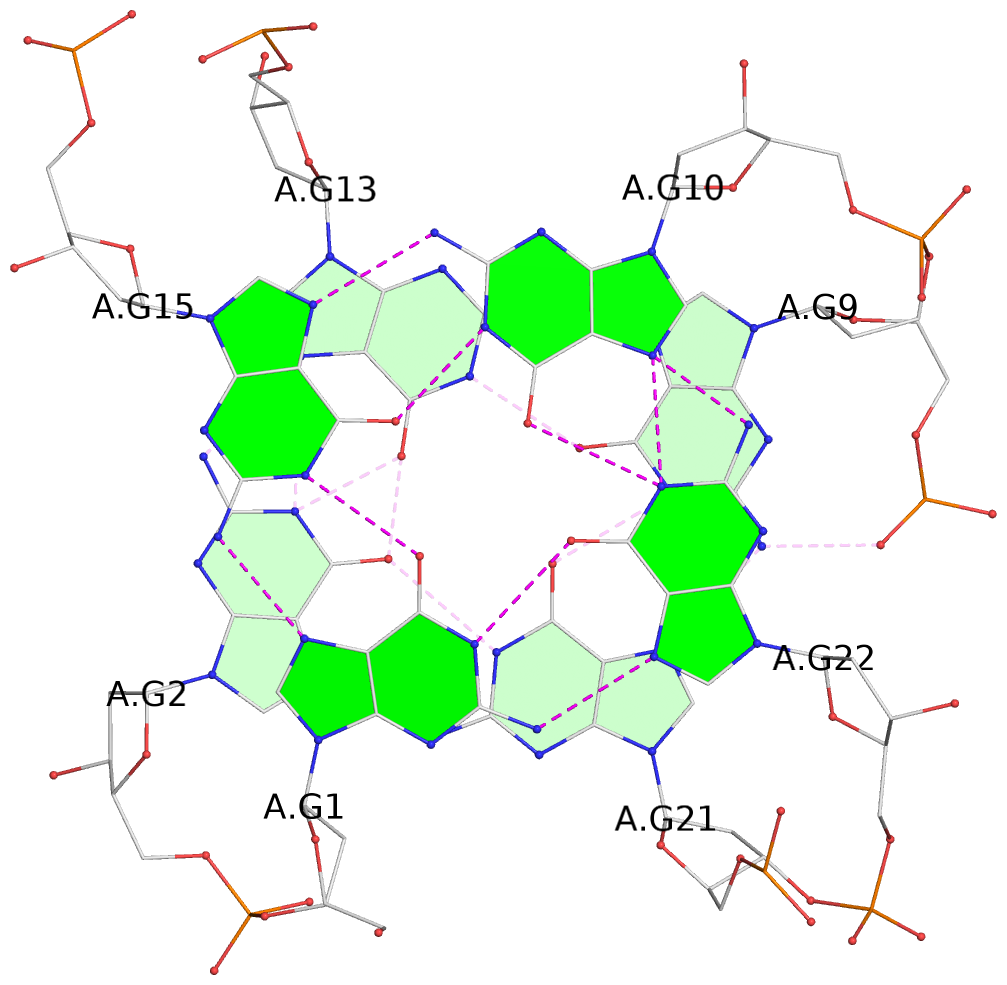
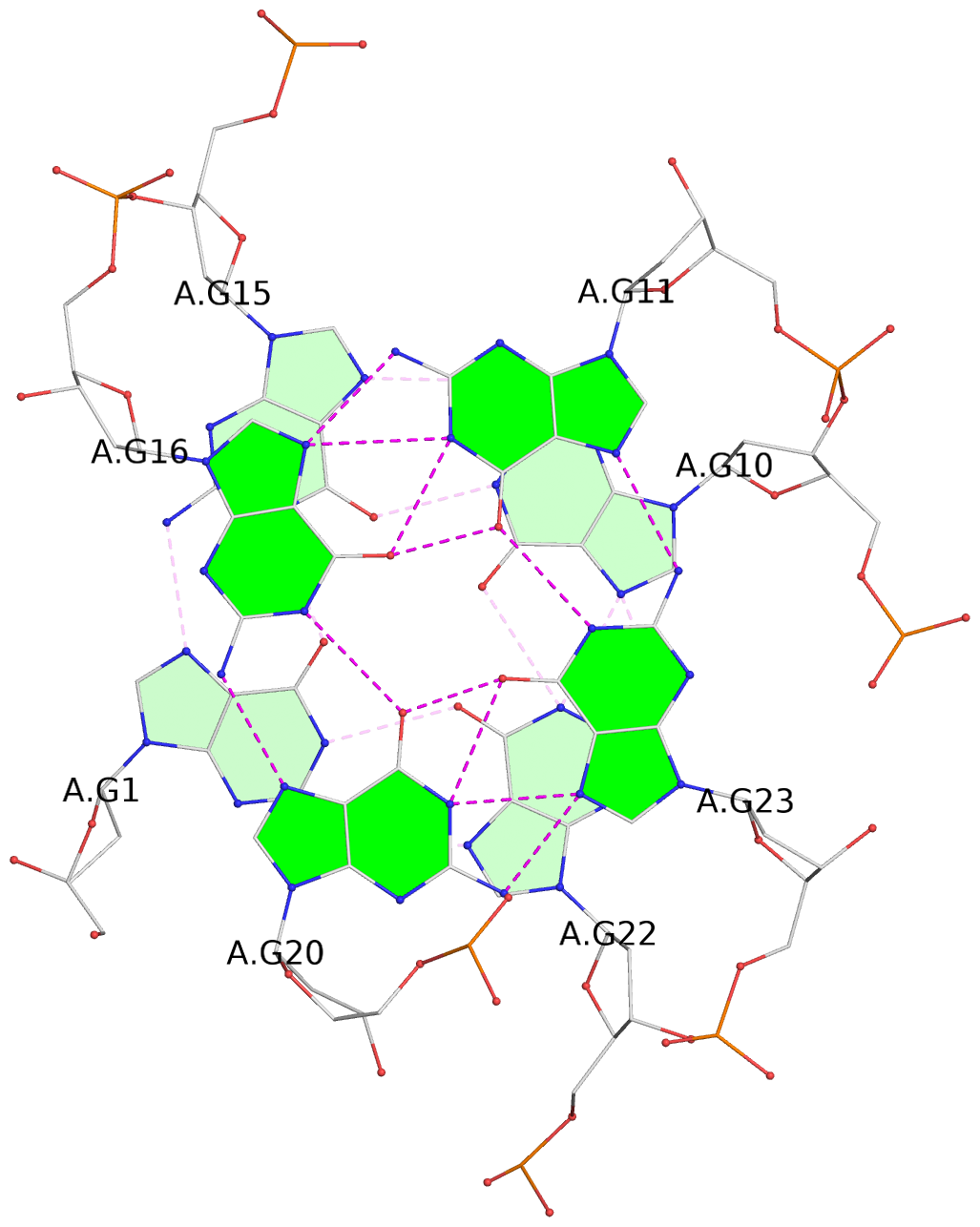
List of 3 G-tetrads
1 glyco-bond=s--- sugar=-... groove=w--n planarity=0.196 type=other nts=4 GGGG A.DG1,A.DG15,A.DG10,A.DG22 2 glyco-bond=--s- sugar=3.-3 groove=-wn- planarity=0.203 type=other nts=4 GGGG A.DG2,A.DG13,A.DG9,A.DG21 3 glyco-bond=--s- sugar=-.3- groove=-wn- planarity=0.185 type=other nts=4 GGGG A.DG11,A.DG16,A.DG20,A.DG23
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UDDD, hybrid-(mixed), 2(D+PX), UD3(1+3)
List of 2 non-stem G4-loops (including the two closing Gs)
1 type=lateral helix=#1 nts=5 GTGCG A.DG16,A.DT17,A.DG18,A.DC19,A.DG20 2 type=V-shaped helix=#1 nts=4 GGGG A.DG20,A.DG21,A.DG22,A.DG23