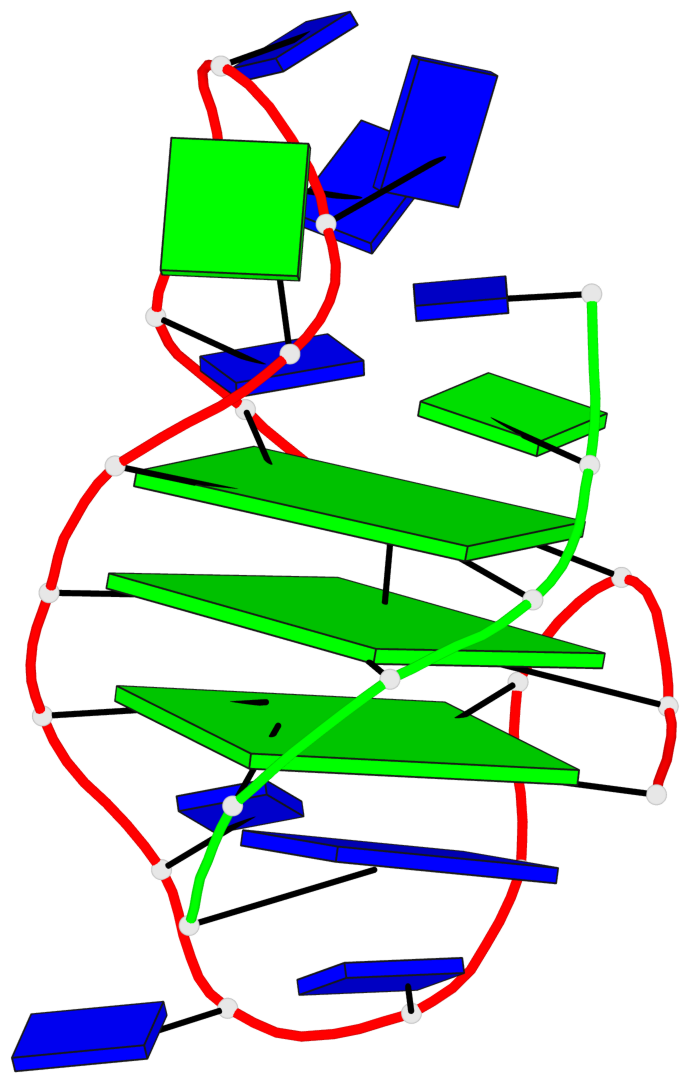
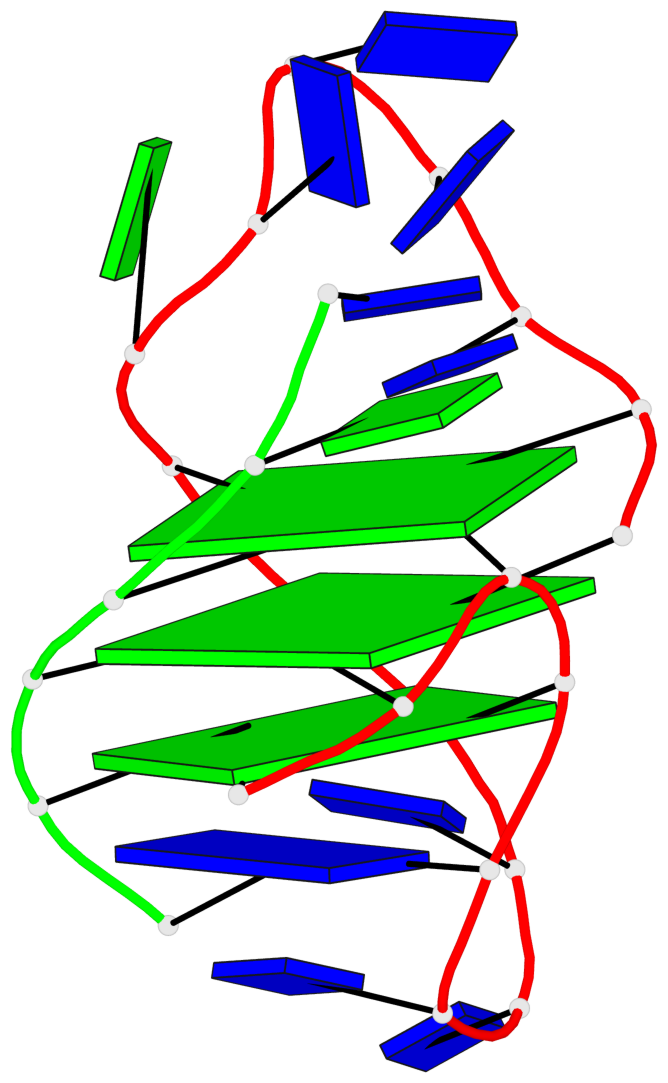
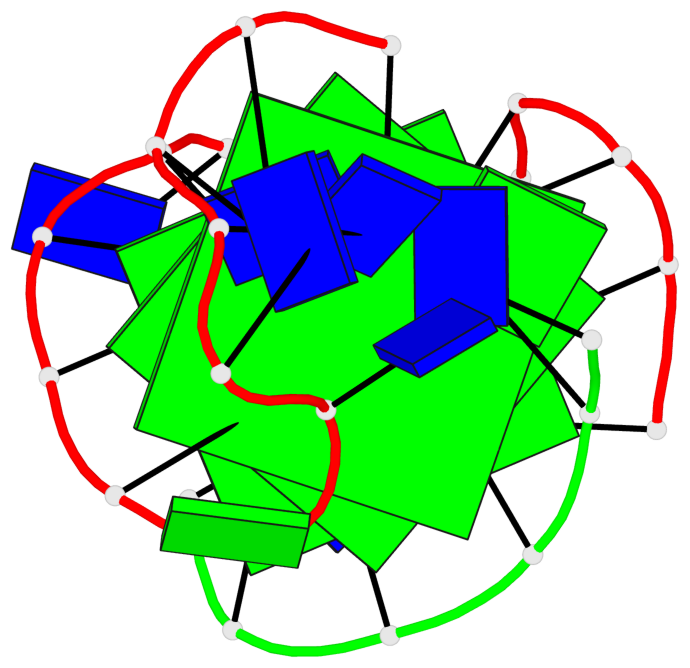
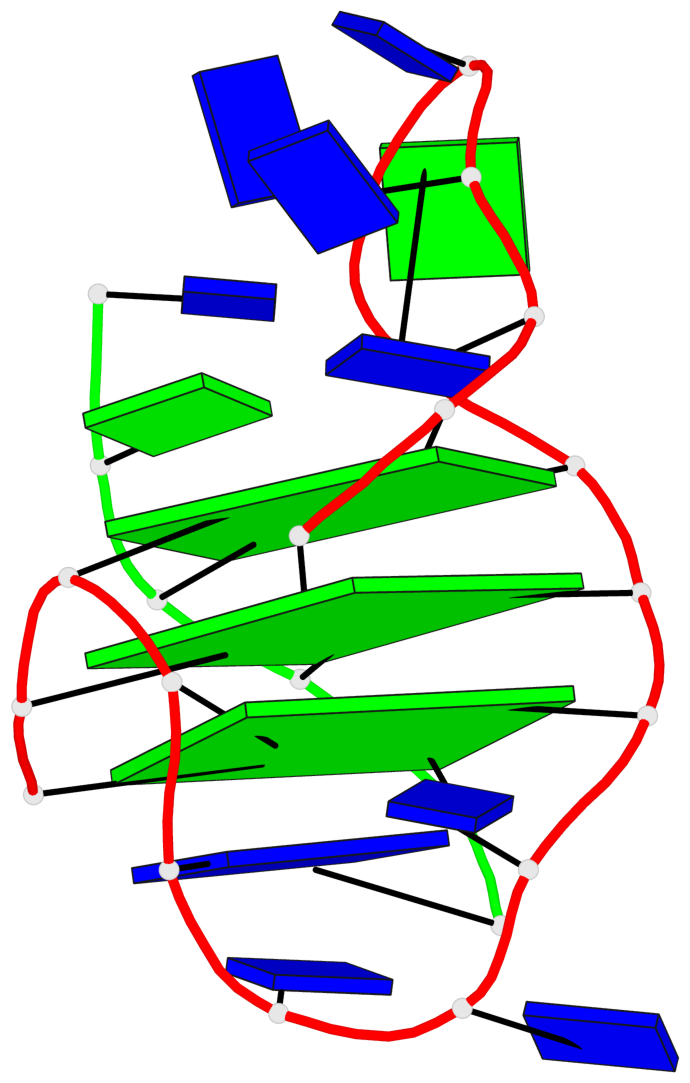
Detailed DSSR results for the G-quadruplex: PDB entry 6a7y
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6a7y
- Class
- DNA
- Method
- NMR
- Summary
- Solution structure of an intermolecular leaped v-shape g-quadruplex
- Reference
- Wan C, Fu W, Jing H, Zhang N (2019): "NMR solution structure of an asymmetric intermolecular leaped V-shape G-quadruplex: selective recognition of the d(G2NG3NG4) sequence motif by a short linear G-rich DNA probe." Nucleic Acids Res., 47, 1544-1556. doi: 10.1093/nar/gky1167.
- Abstract
- Aside from classical loops among G-quadruplexes, the unique leaped V-shape scaffold spans over three G-tetrads, without any intervening residues. This scaffold enables a sharp reversal of two adjacent strand directions and simultaneously participates in forming the G-tetrad core. These features make this scaffold itself distinctive and thus an essentially more accessible target. As an alternative to the conventional antisense method using a complementary chain, forming an intermolecular G-quadruplex from two different oligomers, in which the longer one as the target is captured by a short G-rich fragment, could be helpful for recognizing G-rich sequences and structural motifs. However, such an intermolecular leaped V-shape G-quadruplex consisting of DNA oligomers of quite different lengths has not been evaluated. Here, we present the first nuclear magnetic resonance (NMR) study of an asymmetric intermolecular leaped V-shape G-quadruplex assembled between an Oxytricha nova telomeric sequence d(G2T4G4T4G4) and a single G-tract fragment d(TG4A). Furthermore, we explored the selectivity of this short fragment as a potential probe, examined the kinetic discrimination for probing a specific mutant, and proposed the key sequence motif d(G2NG3NG4) essential for building the leaped V-shape G-quadruplexes.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, (1+3), UDDD
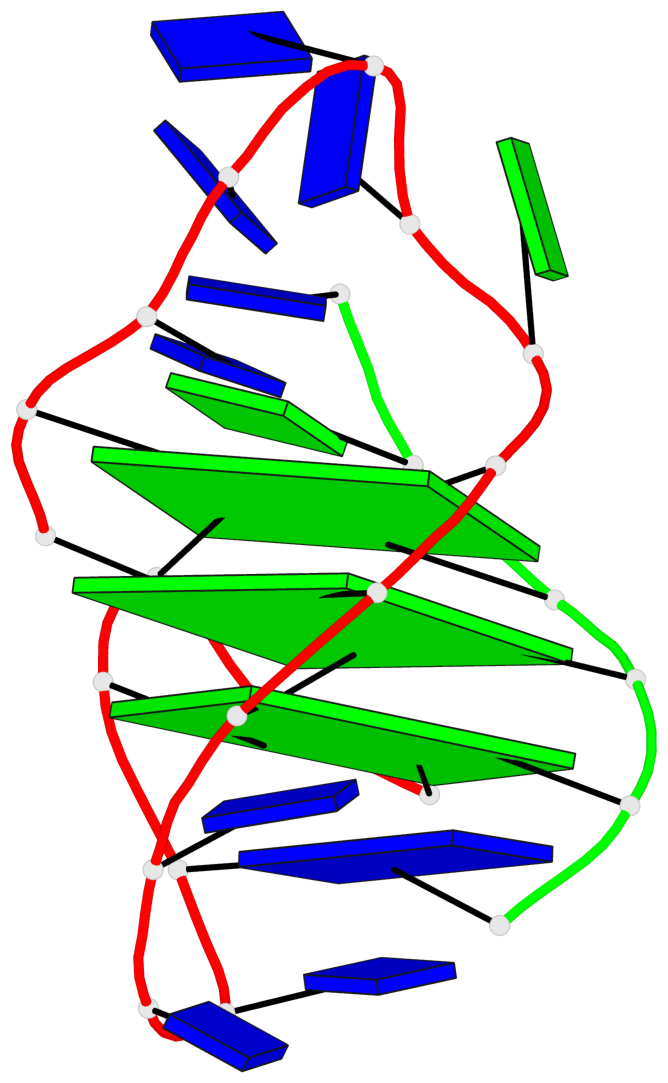
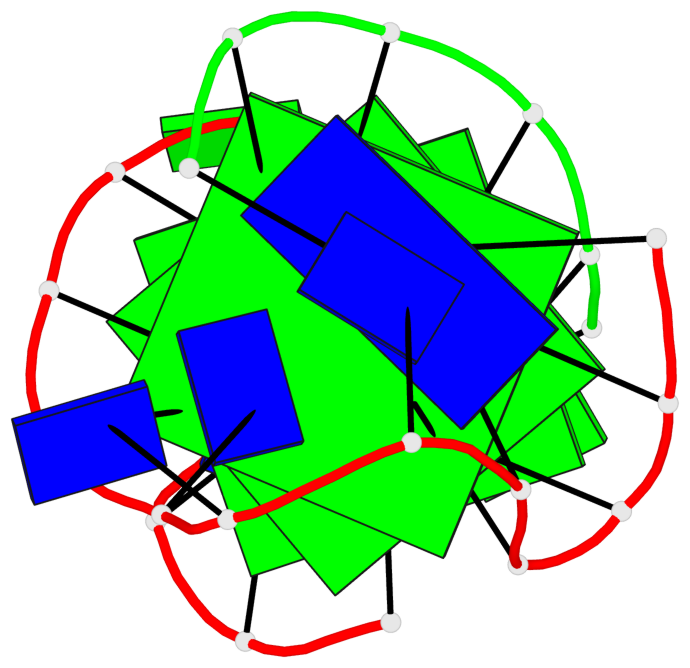
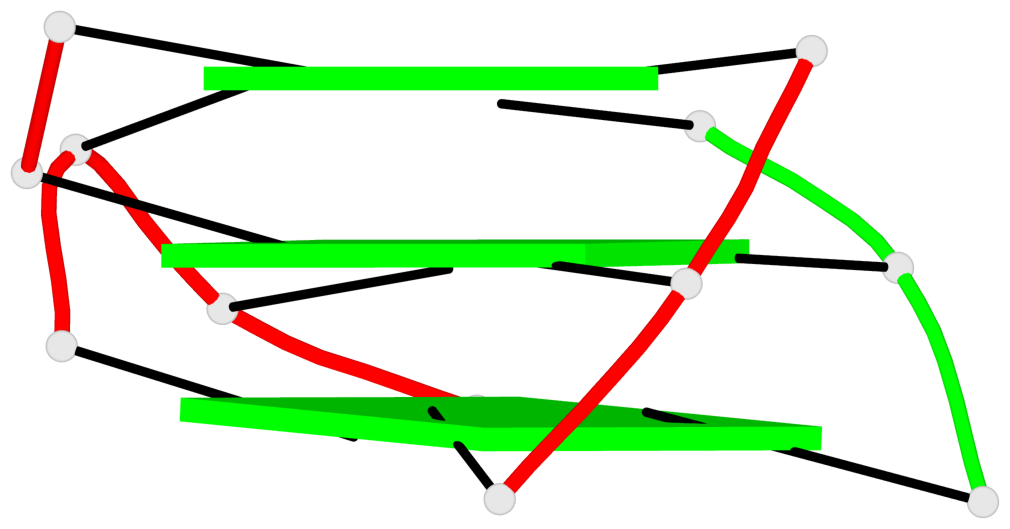
Base-block schematics in six views
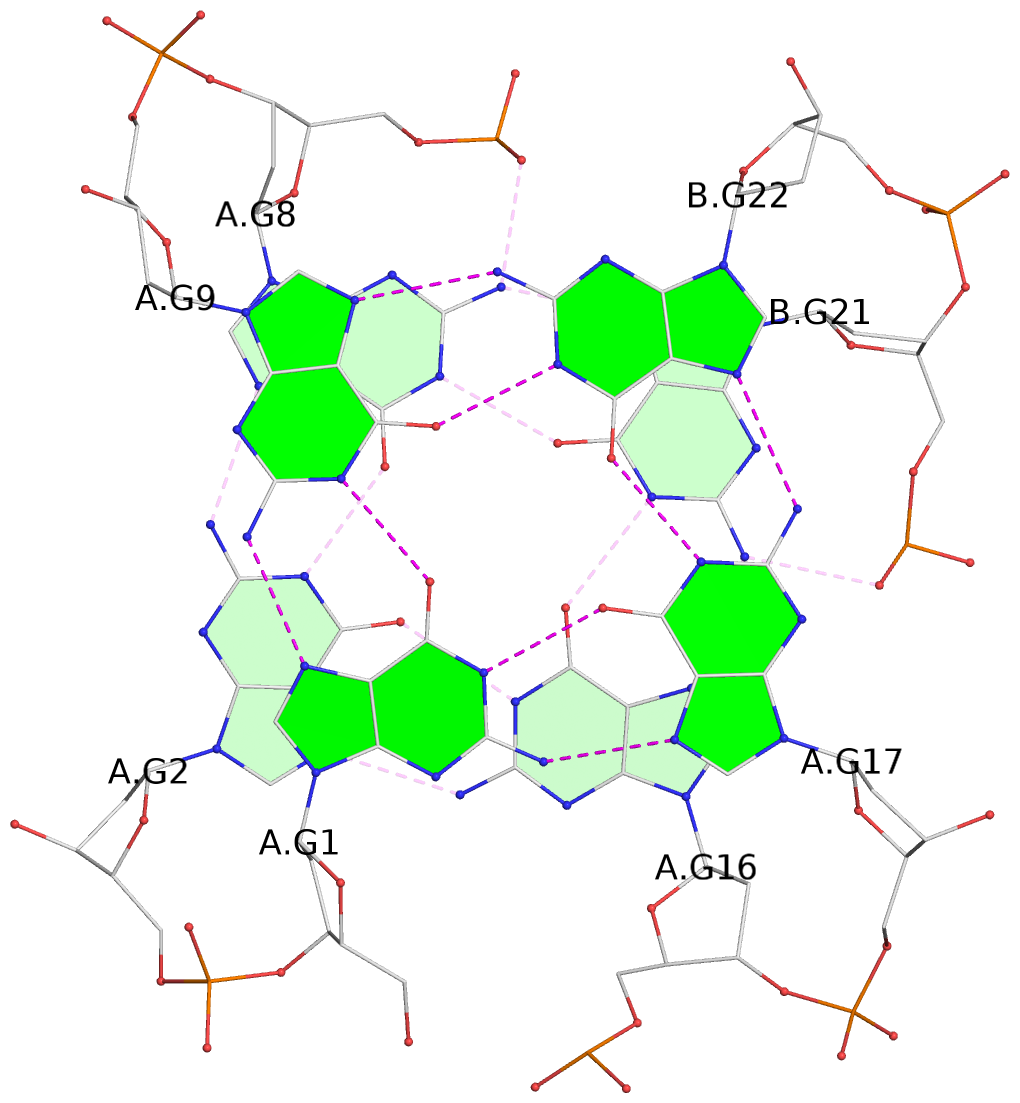
List of 3 G-tetrads
1 glyco-bond=s--- sugar=.3-- groove=w--n planarity=0.243 type=other nts=4 GGGG A.DG1,A.DG9,B.DG22,A.DG17 2 glyco-bond=-sss sugar=---- groove=w--n planarity=0.334 type=other nts=4 GGGG A.DG2,A.DG8,B.DG21,A.DG16 3 glyco-bond=-s-- sugar=---- groove=wn-- planarity=0.322 type=other nts=4 GGGG A.DG10,A.DG15,A.DG18,B.DG23
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 2 loops, inter-molecular, UDDD, hybrid-(mixed), (1+3)
List of 2 non-stem G4-loops (including the two closing Gs)
1 type=lateral helix=#1 nts=6 GTTTTG A.DG10,A.DT11,A.DT12,A.DT13,A.DT14,A.DG15 2 type=V-shaped helix=#1 nts=4 GGGG A.DG15,A.DG16,A.DG17,A.DG18