Detailed DSSR results for the G-quadruplex: PDB entry 6a85
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6a85
- Class
- DNA
- Method
- X-ray (1.45 Å)
- Summary
- Crystal structure of a novel DNA quadruplex
- Reference
- Liu HH, Wang R, Yu X, Shen FS, Lan WX, Haruehanroengra P, Yao QQ, Zhang J, Chen YQ, Li SH, Wu BX, Zheng LN, Ma JB, Lin JZ, Cao CY, Li JX, Sheng J, Gan JH (2018): "High-resolution DNA quadruplex structure containing all the A-, G-, C-, T-tetrads." Nucleic Acids Res., 46, 11627-11638. doi: 10.1093/nar/gky902.
- Abstract
- DNA can form diverse structures, which predefine their physiological functions. Besides duplexes that carry the genetic information, quadruplexes are the most well-studied DNA structures. In addition to their important roles in recombination, replication, transcription and translation, DNA quadruplexes have also been applied as diagnostic aptamers and antidisease therapeutics. Herein we further expand the sequence and structure complexity of DNA quadruplex by presenting a high-resolution crystal structure of DNA1 (5'-AGAGAGATGGGTGCGTT-3'). This is the first quadruplex structure that contains all the internal A-, G-, C-, T-tetrads, A:T:A:T tetrads and bulged nucleotides in one single structure; as revealed by site-specific mutagenesis and biophysical studies, the central ATGGG motif plays important role in the quadruplex formation. Interestingly, our structure also provides great new insights into cation recognition, including the first-time reported Pb2+, by tetrad structures.
- G4 notes
- 8 G-tetrads, 2 G4 helices, 1 G4 stem, parallel(4+0), UUUU
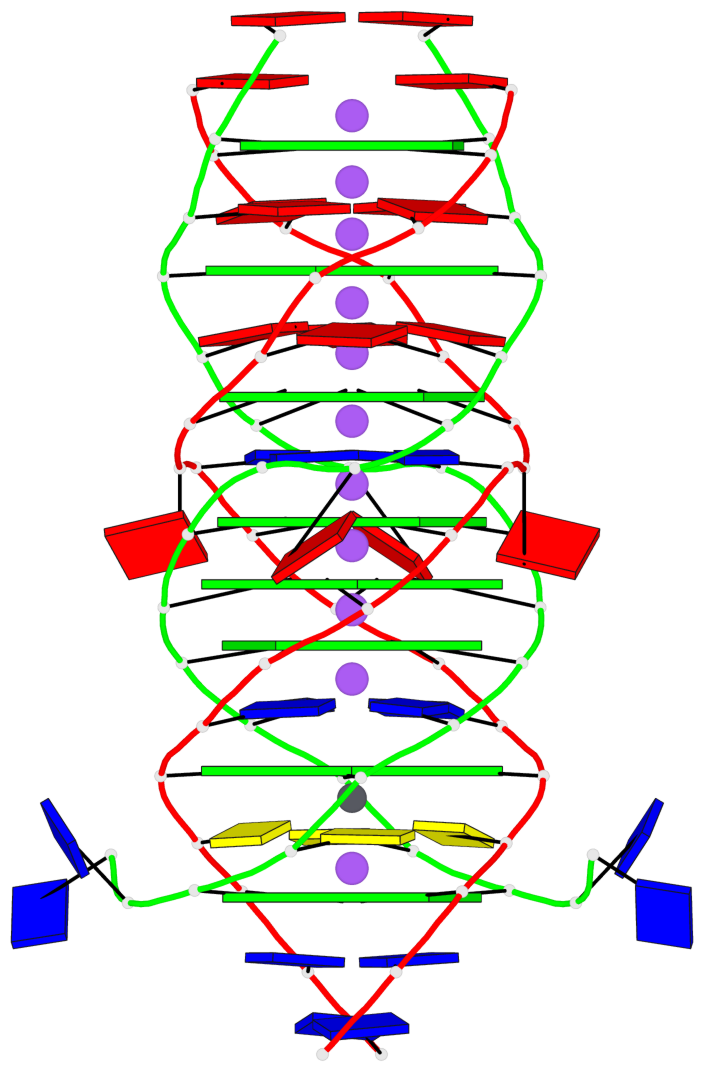
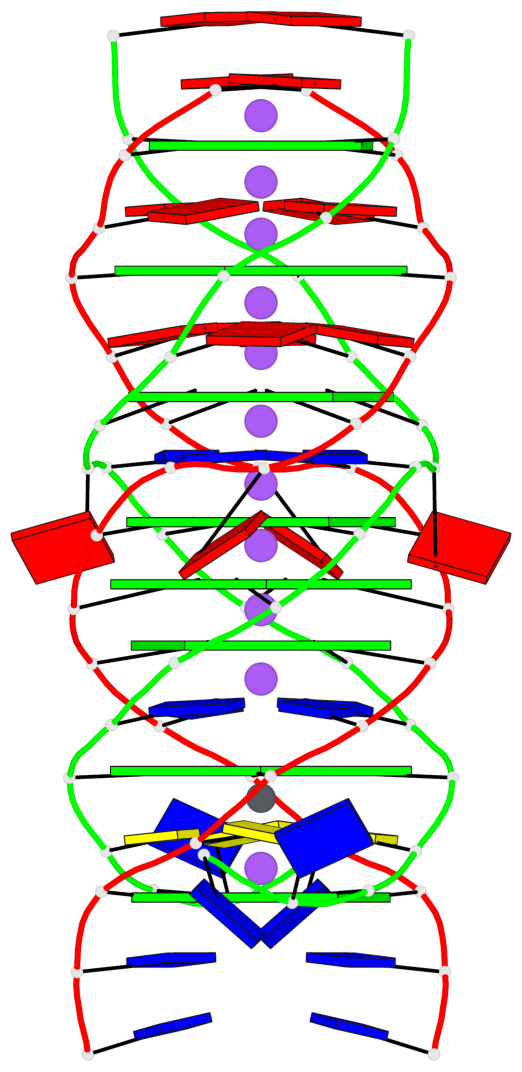
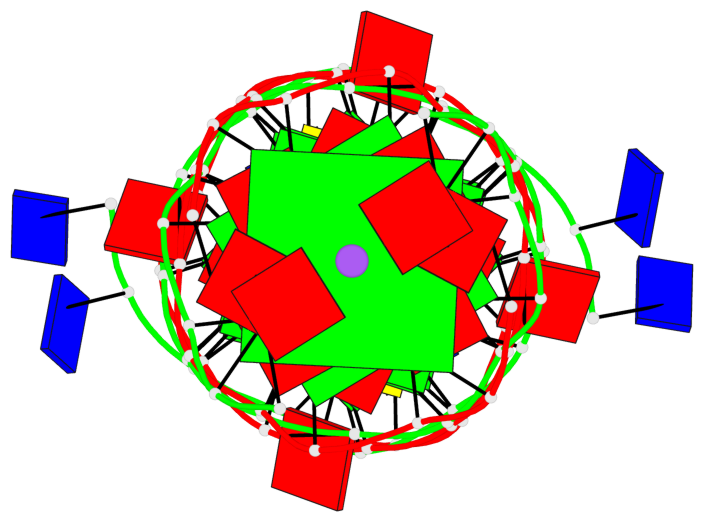
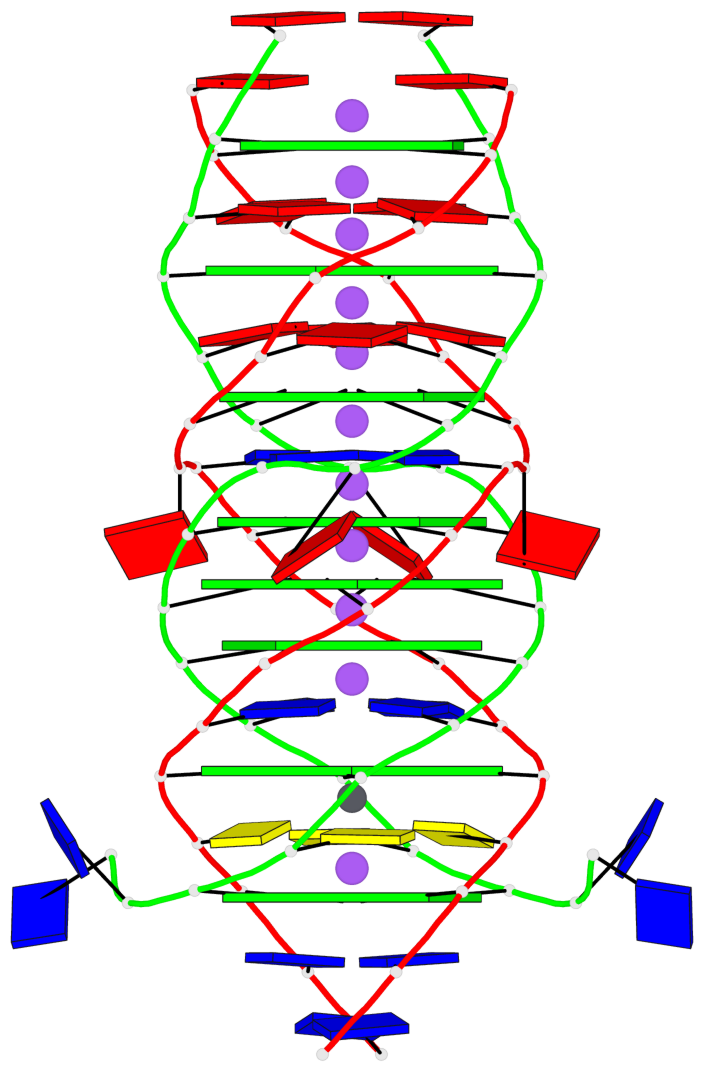
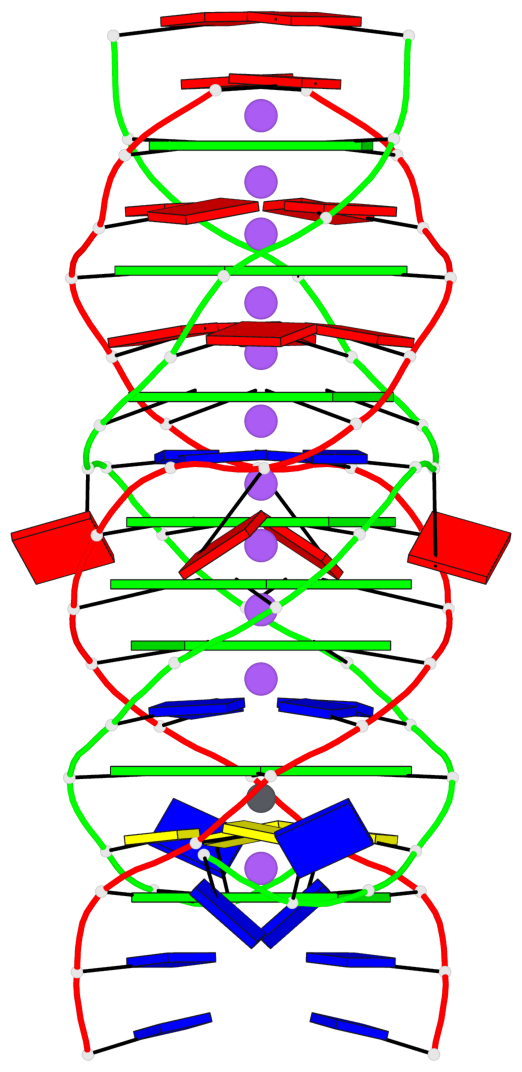
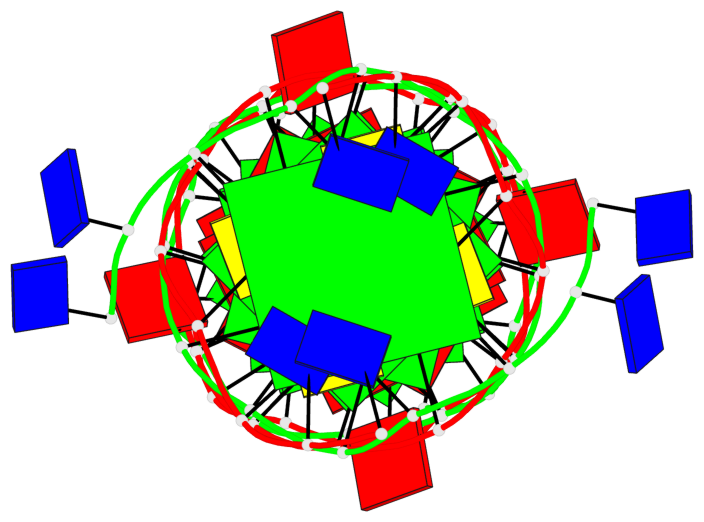
Base-block schematics in six views
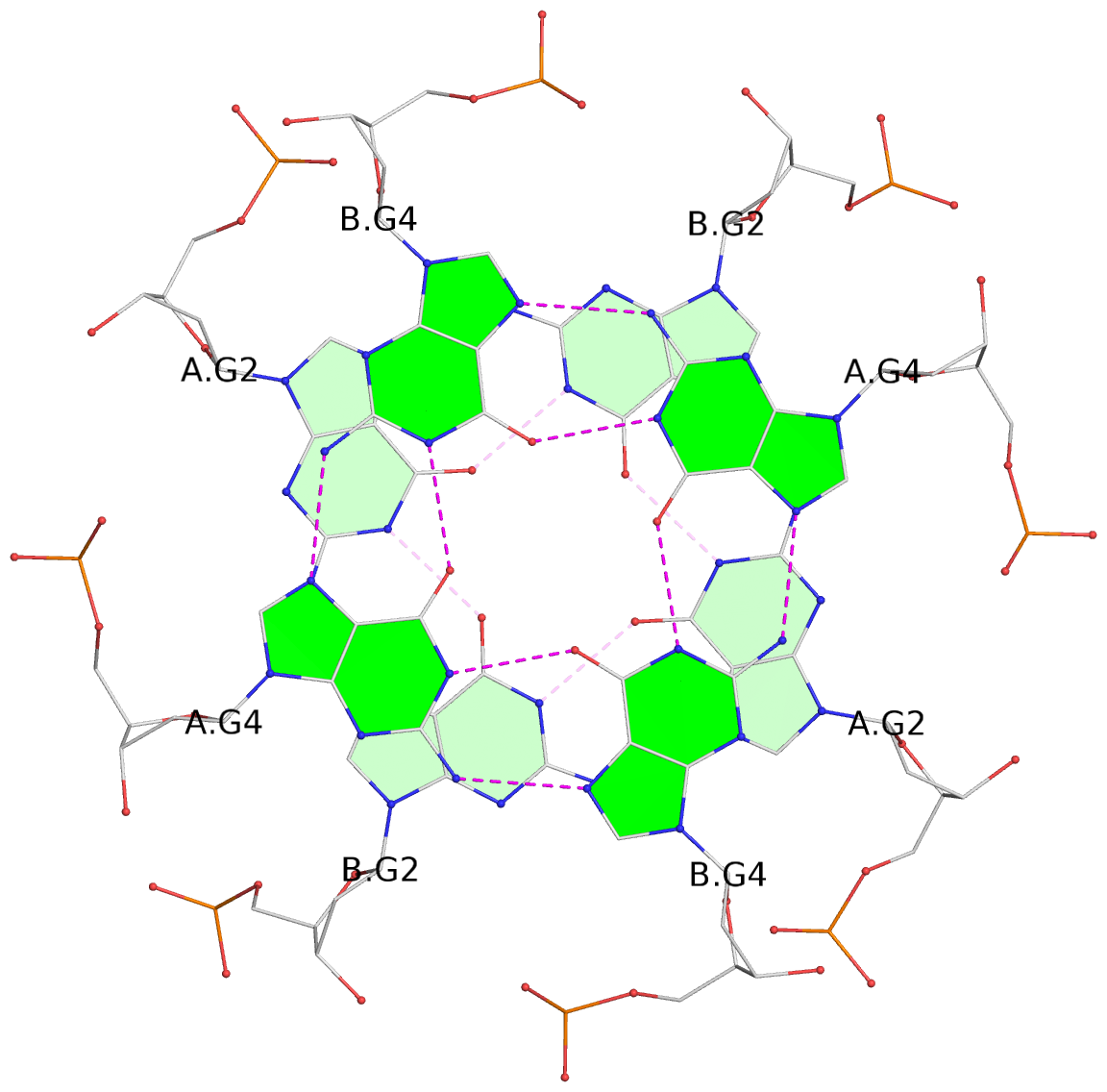
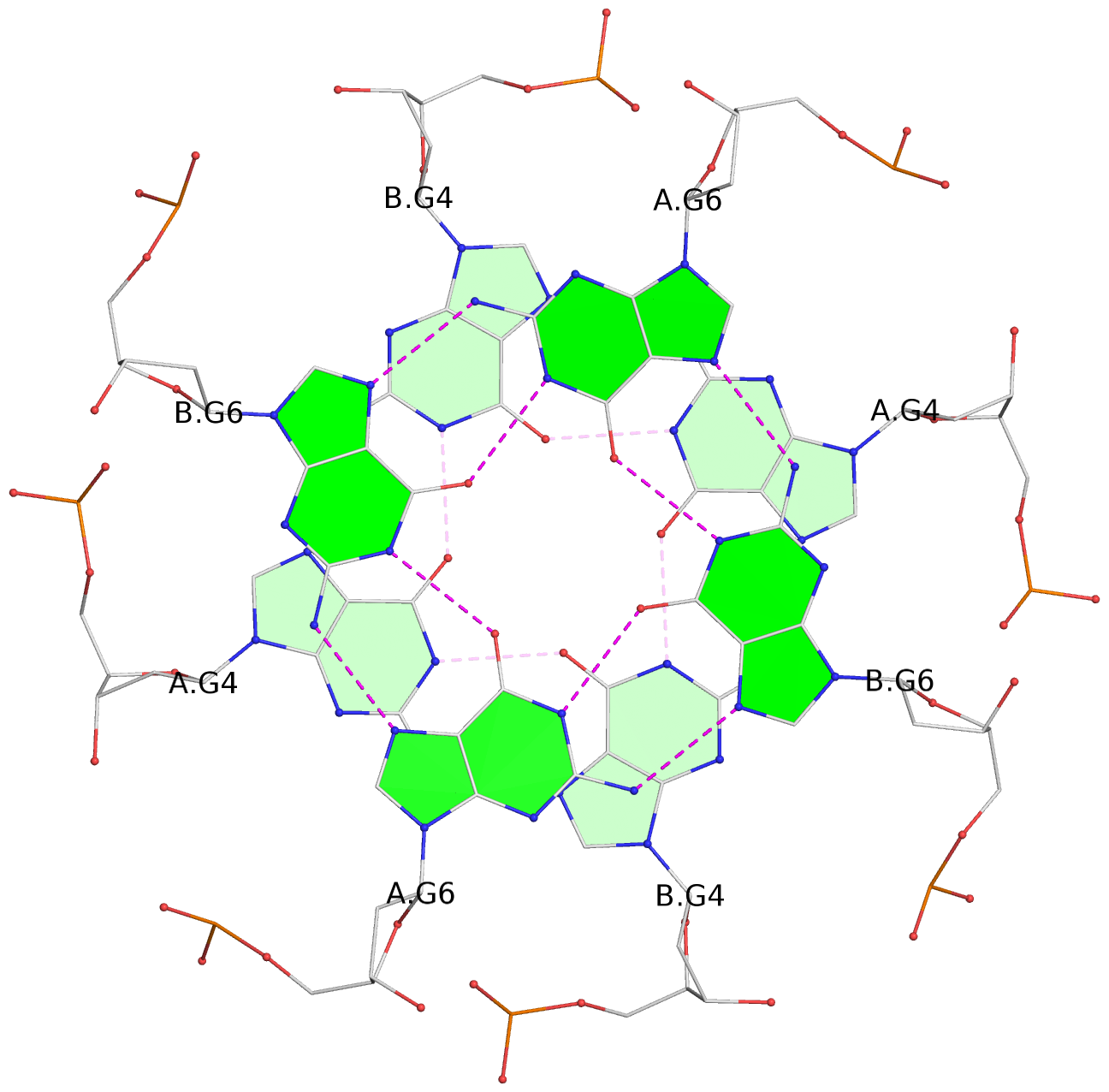
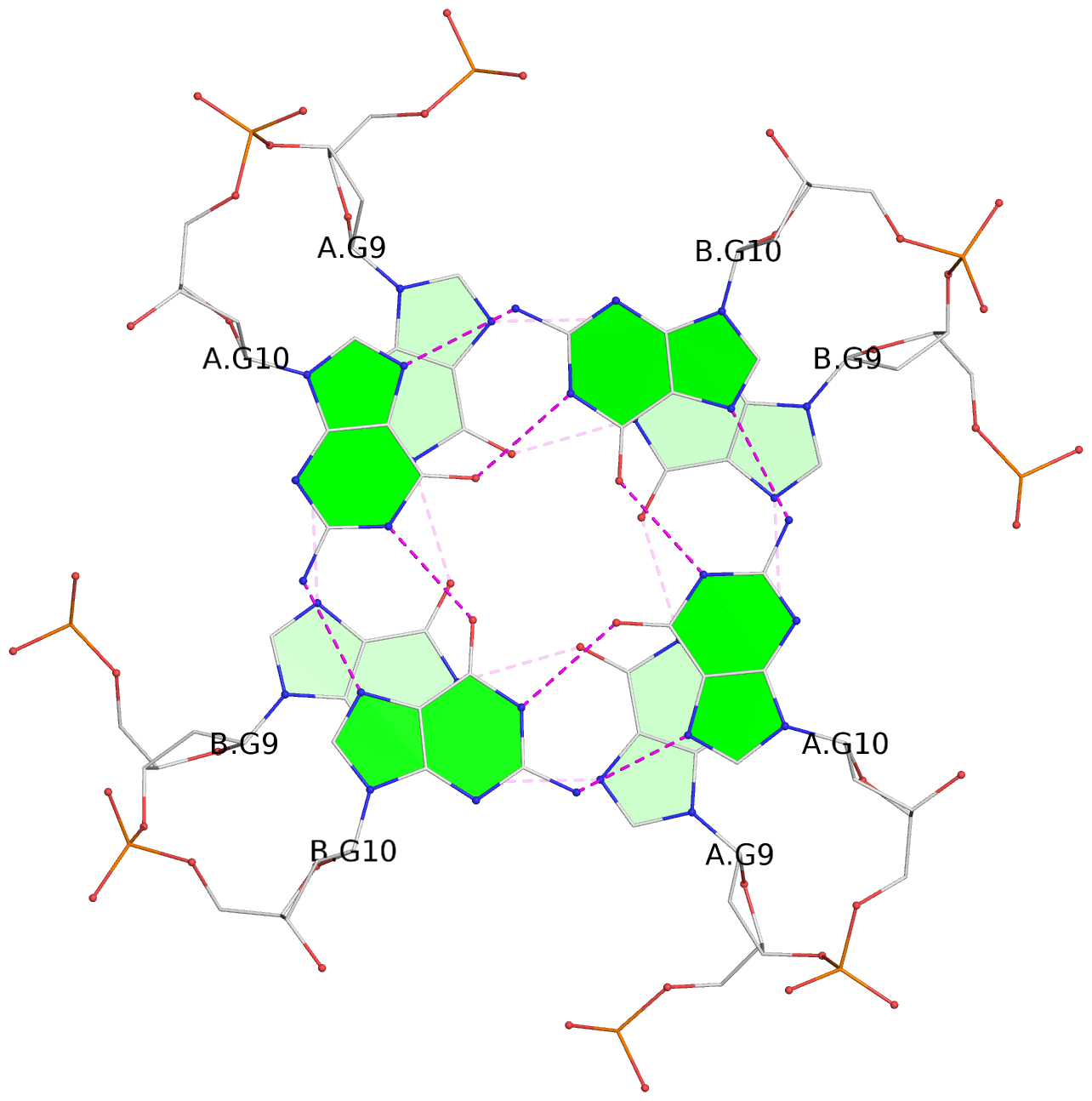
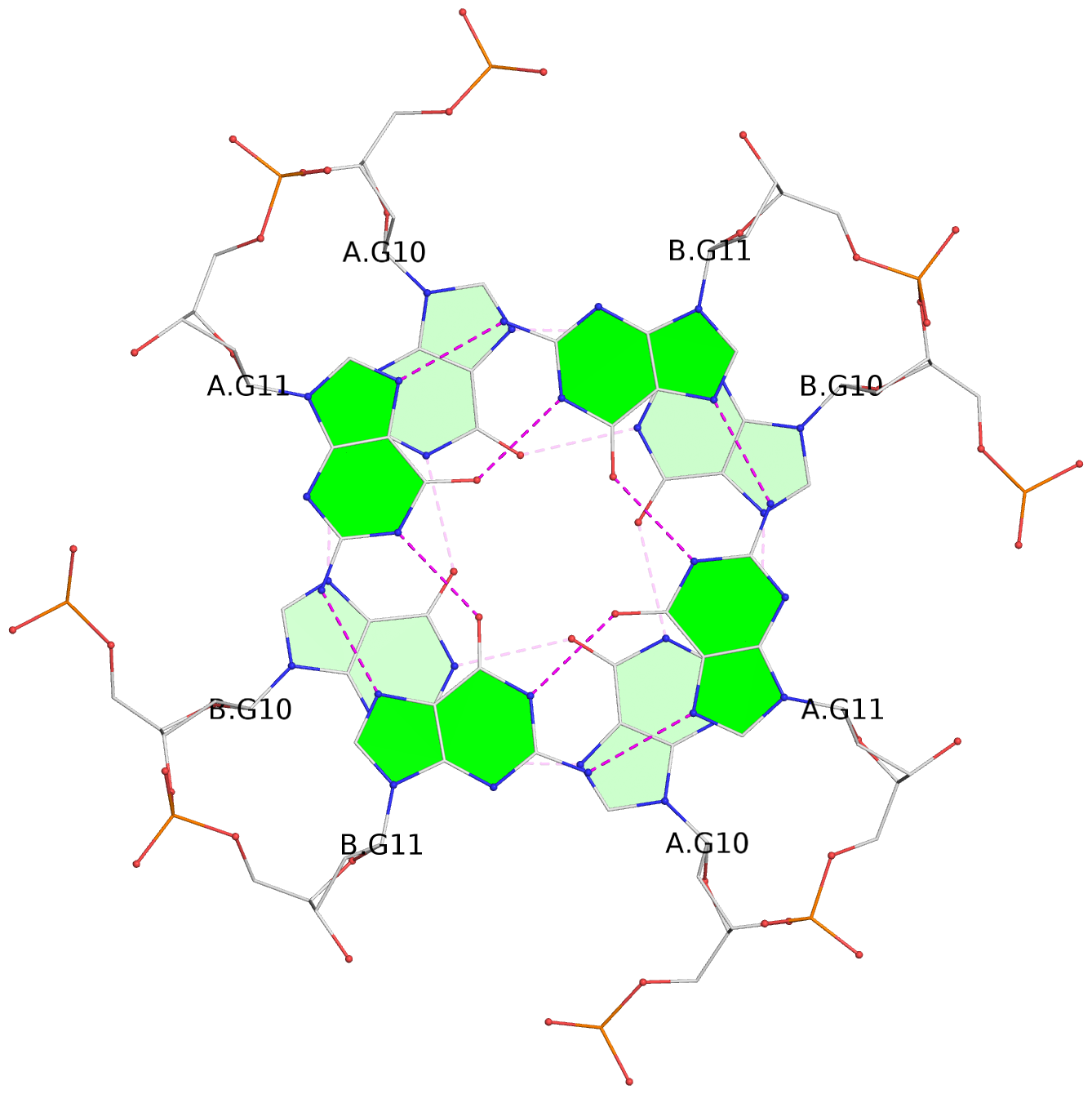
List of 8 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.208 type=other nts=4 GGGG 1:A.DG2,1:B.DG2,2:A.DG2,2:B.DG2 2 glyco-bond=---- sugar=---- groove=---- planarity=0.107 type=planar nts=4 GGGG 1:A.DG4,1:B.DG4,2:A.DG4,2:B.DG4 3 glyco-bond=---- sugar=---- groove=---- planarity=0.300 type=bowl nts=4 GGGG 1:A.DG6,1:B.DG6,2:A.DG6,2:B.DG6 4 glyco-bond=---- sugar=---- groove=---- planarity=0.191 type=other nts=4 GGGG 1:A.DG9,1:B.DG9,2:A.DG9,2:B.DG9 5 glyco-bond=---- sugar=---- groove=---- planarity=0.219 type=bowl nts=4 GGGG 1:A.DG10,1:B.DG10,2:A.DG10,2:B.DG10 6 glyco-bond=---- sugar=---- groove=---- planarity=0.159 type=planar nts=4 GGGG 1:A.DG11,1:B.DG11,2:A.DG11,2:B.DG11 7 glyco-bond=---- sugar=---- groove=---- planarity=0.091 type=planar nts=4 GGGG 1:A.DG13,1:B.DG13,2:A.DG13,2:B.DG13 8 glyco-bond=---- sugar=---- groove=---- planarity=0.174 type=saddle nts=4 GGGG 1:A.DG15,1:B.DG15,2:A.DG15,2:B.DG15
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular
Helix#2, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.