Detailed DSSR results for the G-quadruplex: PDB entry 6au4
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6au4
- Class
- DNA
- Method
- X-ray (2.35 Å)
- Summary
- Crystal structure of the major quadruplex formed in the human c-myc promoter
- Reference
- Stump S, Mou TC, Sprang SR, Natale NR, Beall HD (2018): "Crystal structure of the major quadruplex formed in the promoter region of the human c-MYC oncogene." PLoS ONE, 13, e0205584. doi: 10.1371/journal.pone.0205584.
- Abstract
- The c-MYC oncogene mediates multiple tumor cell survival pathways and is dysregulated or overexpressed in the majority of human cancers. The NHE III1 region of the c-MYC promoter forms a DNA quadruplex. Stabilization of this structure with small molecules has been shown to reduce expression of c-MYC, and targeting the c-MYC quadruplex has become an emerging strategy for development of antitumor compounds. Previous solution NMR studies of the c-MYC quadruplex have assigned the major conformer and topology of this important target, however, regions outside the G-quartet core were not as well-defined. Here, we report a high-resolution crystal structure (2.35 Å) of the major quadruplex formed in the NHE III1 region of the c-MYC promoter. The crystal structure is in general agreement with the solution NMR structure, however, key differences are observed in the position of nucleotides outside the G-quartet core. The crystal structure provides an alternative model that, along with comparisons to other reported quadruplex crystal structures, will be important to the rational design of selective compounds. This work will aid in development of ligands to target the c-MYC promoter quadruplex with the goal of creating novel anticancer therapies.
- G4 notes
- 6 G-tetrads, 2 G4 helices, 2 G4 stems, 3(-P-P-P), parallel(4+0), UUUU
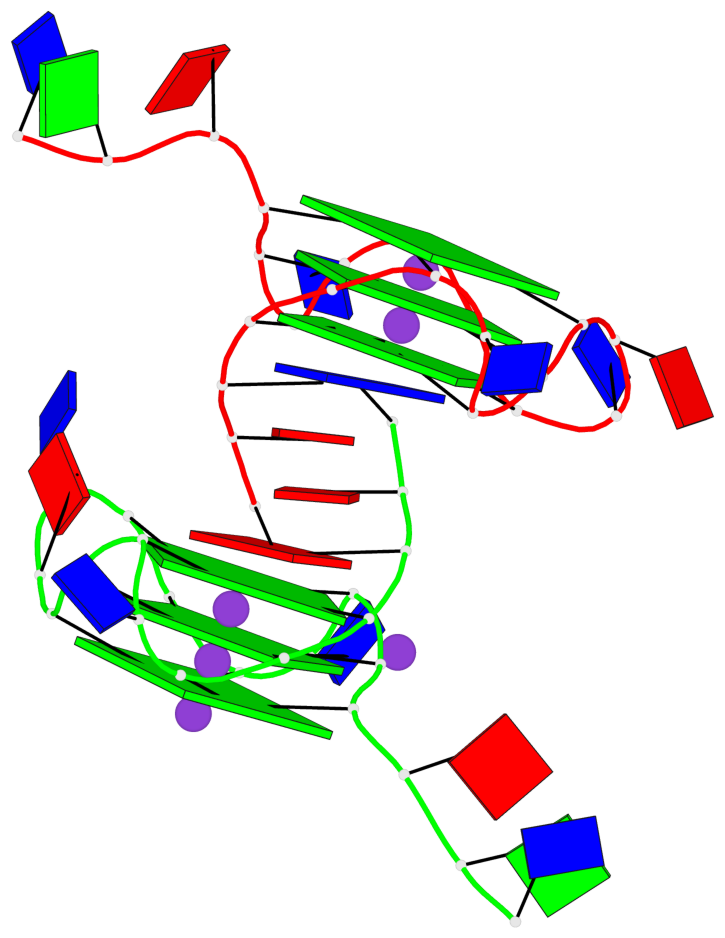
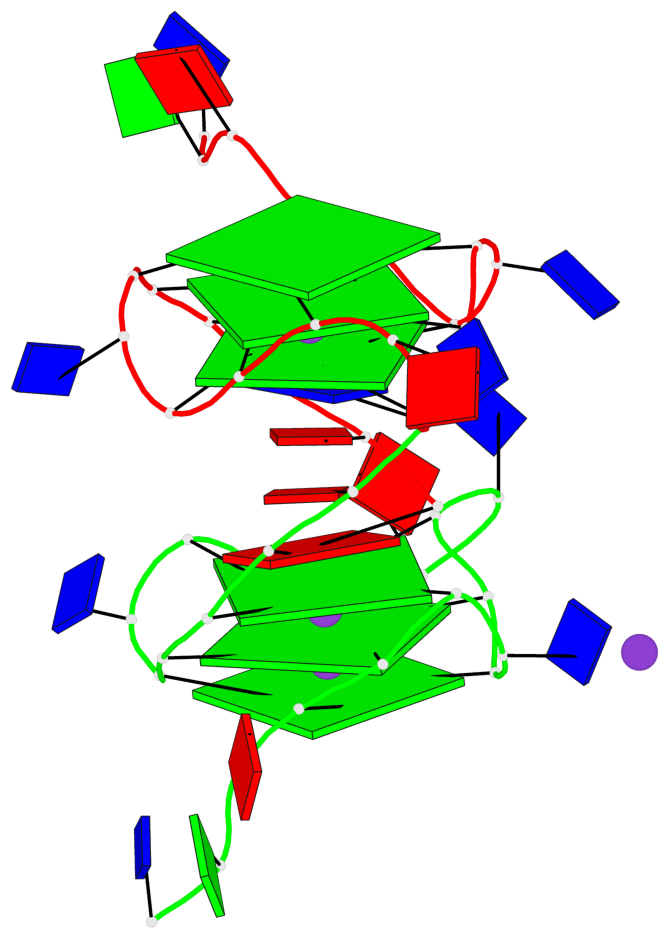
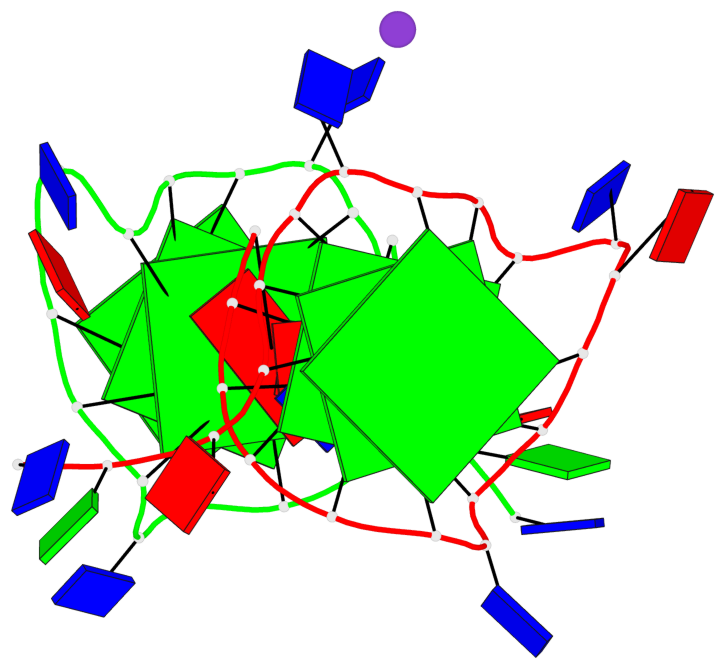
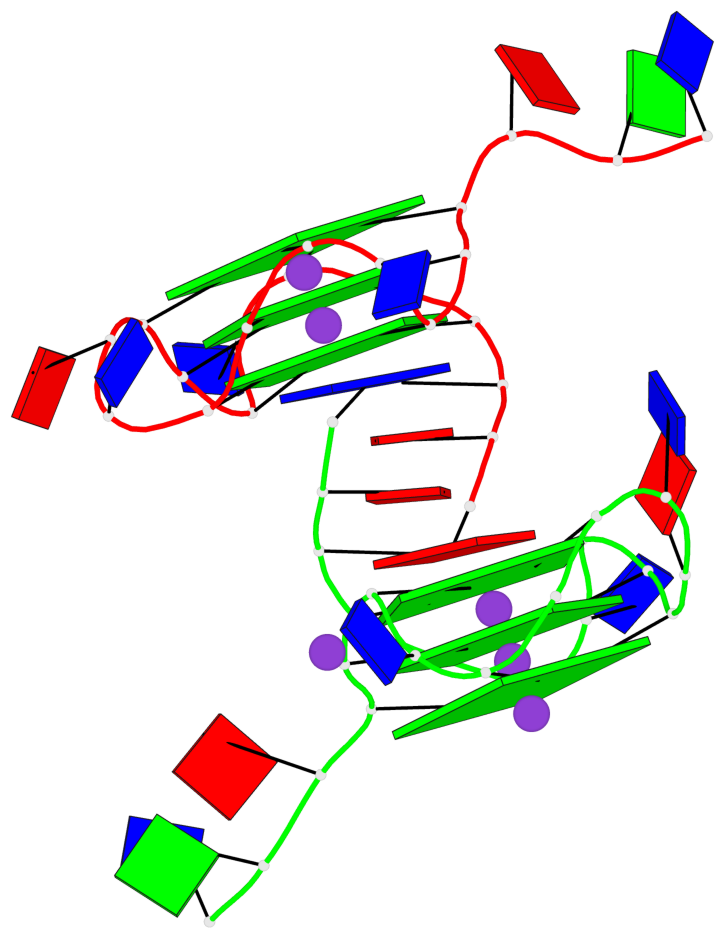
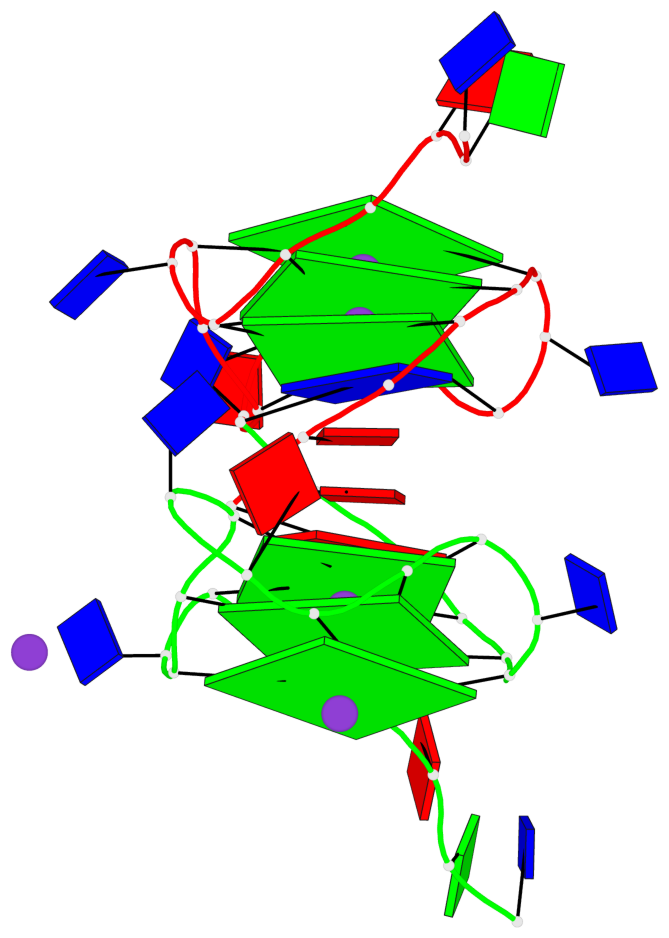
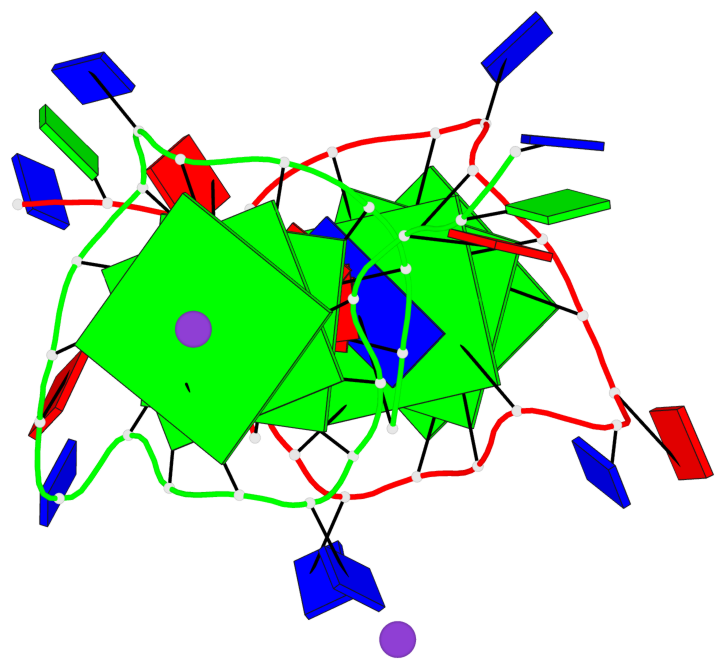
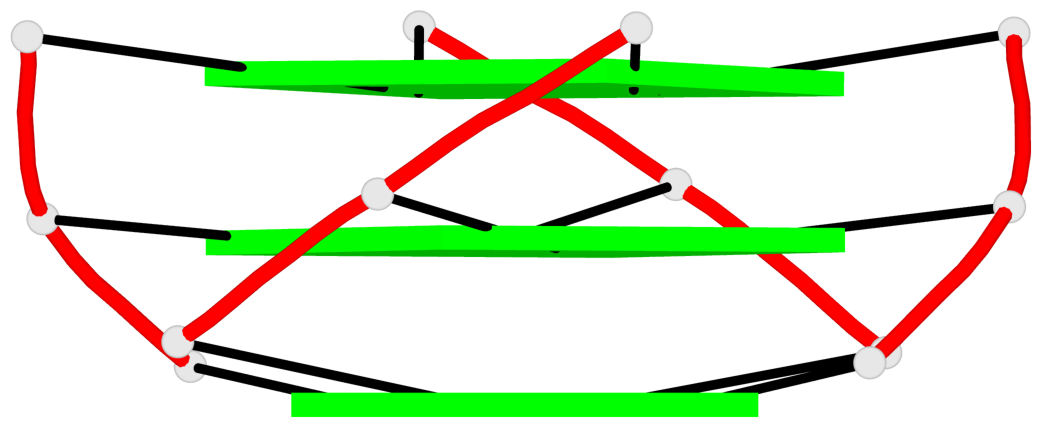
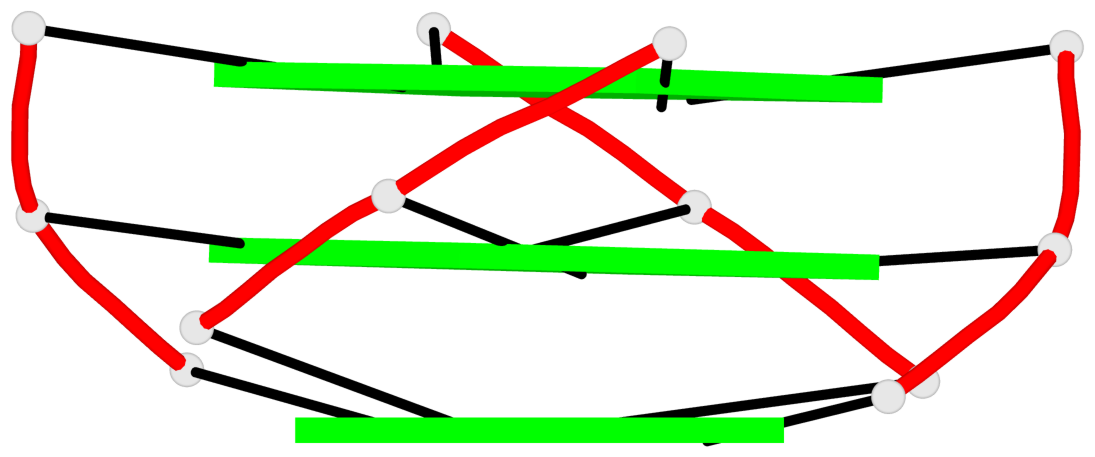
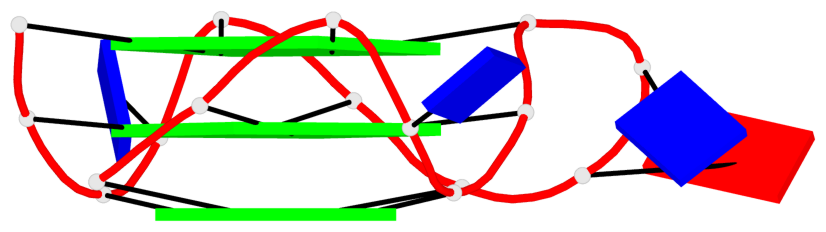
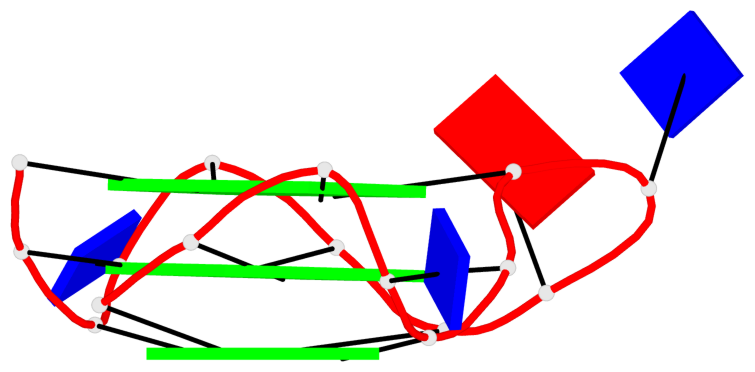
Base-block schematics in six views
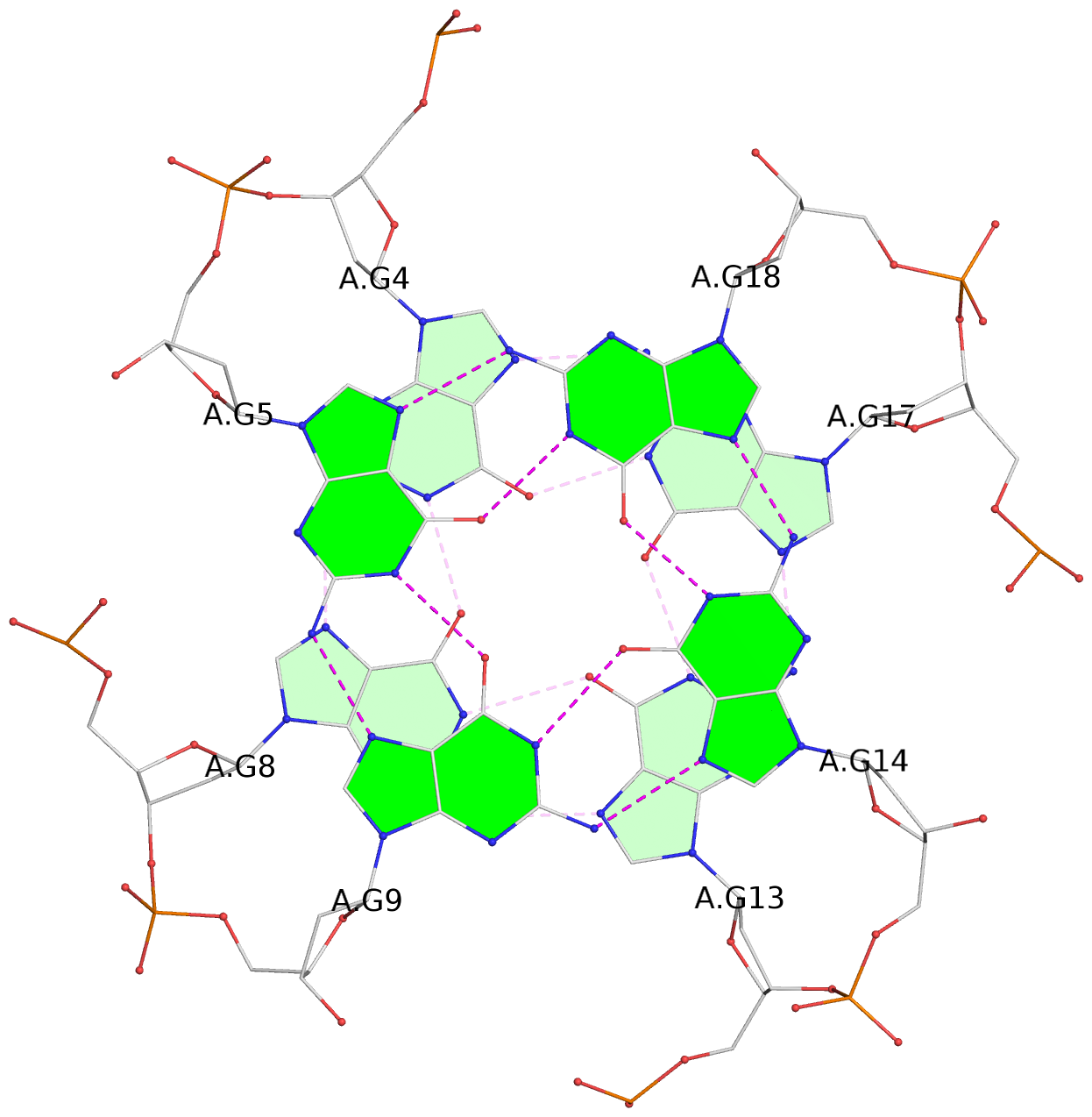
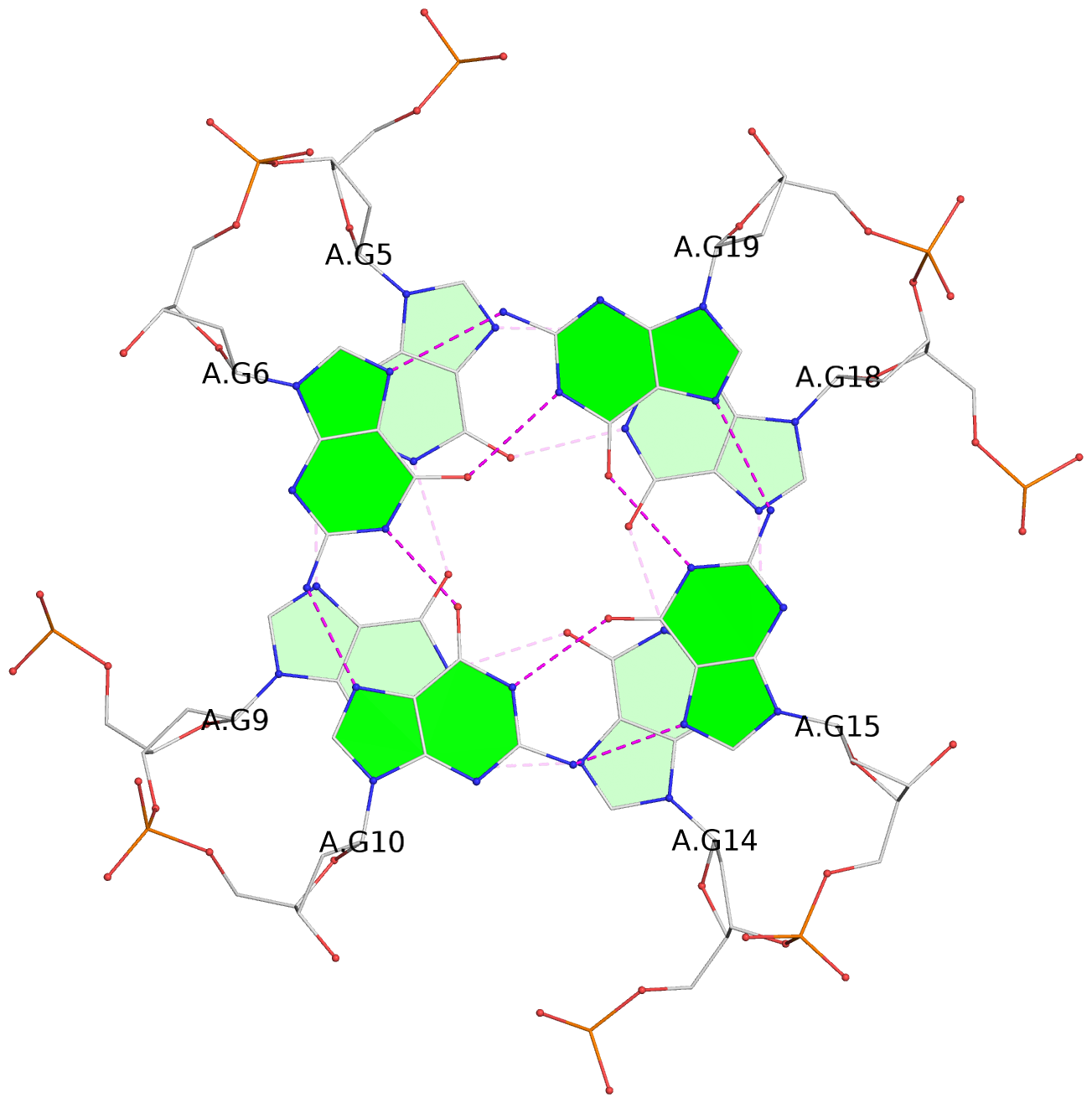
List of 6 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.043 type=planar nts=4 GGGG A.DG4,A.DG8,A.DG13,A.DG17 2 glyco-bond=---- sugar=--.- groove=---- planarity=0.130 type=planar nts=4 GGGG A.DG5,A.DG9,A.DG14,A.DG18 3 glyco-bond=---- sugar=---- groove=---- planarity=0.197 type=bowl nts=4 GGGG A.DG6,A.DG10,A.DG15,A.DG19 4 glyco-bond=---- sugar=3--- groove=---- planarity=0.074 type=planar nts=4 GGGG B.DG4,B.DG8,B.DG13,B.DG17 5 glyco-bond=---- sugar=---- groove=---- planarity=0.140 type=planar nts=4 GGGG B.DG5,B.DG9,B.DG14,B.DG18 6 glyco-bond=---- sugar=---- groove=---- planarity=0.249 type=bowl nts=4 GGGG B.DG6,B.DG10,B.DG15,B.DG19
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
Helix#2, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.