Detailed DSSR results for the G-quadruplex: PDB entry 6ccw
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6ccw
- Class
- DNA
- Method
- NMR
- Summary
- Hybrid-2 form human telomeric g quadruplex in complex with epiberberine
- Reference
- Lin C, Wu G, Wang K, Onel B, Sakai S, Shao Y, Yang D (2018): "Molecular Recognition of the Hybrid-2 Human Telomeric G-Quadruplex by Epiberberine: Insights into Conversion of Telomeric G-Quadruplex Structures." Angew. Chem. Int. Ed. Engl., 57, 10888-10893. doi: 10.1002/anie.201804667.
- Abstract
- Human telomeres can form DNA G-quadruplex (G4), an attractive target for anticancer drugs. Human telomeric G4s bear inherent structure polymorphism, challenging for understanding specific recognition by ligands or proteins. Protoberberines are medicinal natural-products known to stabilize telomeric G4s and inhibit telomerase. Here we report epiberberine (EPI) specifically recognizes the hybrid-2 telomeric G4 predominant in physiologically relevant K+ solution and converts other telomeric G4 forms to hybrid-2, the first such example reported. Our NMR structure in K+ solution shows EPI binding induces extensive rearrangement of the previously disordered 5'-flanking and loop segments to form an unprecedented four-layer binding pocket specific to the hybrid-2 telomeric G4; EPI recruits the (-1) adenine to form a "quasi-triad" intercalated between the external tetrad and a T:T:A triad, capped by a T:T base pair. Our study provides structural basis for small-molecule drug design targeting the human telomeric G4.
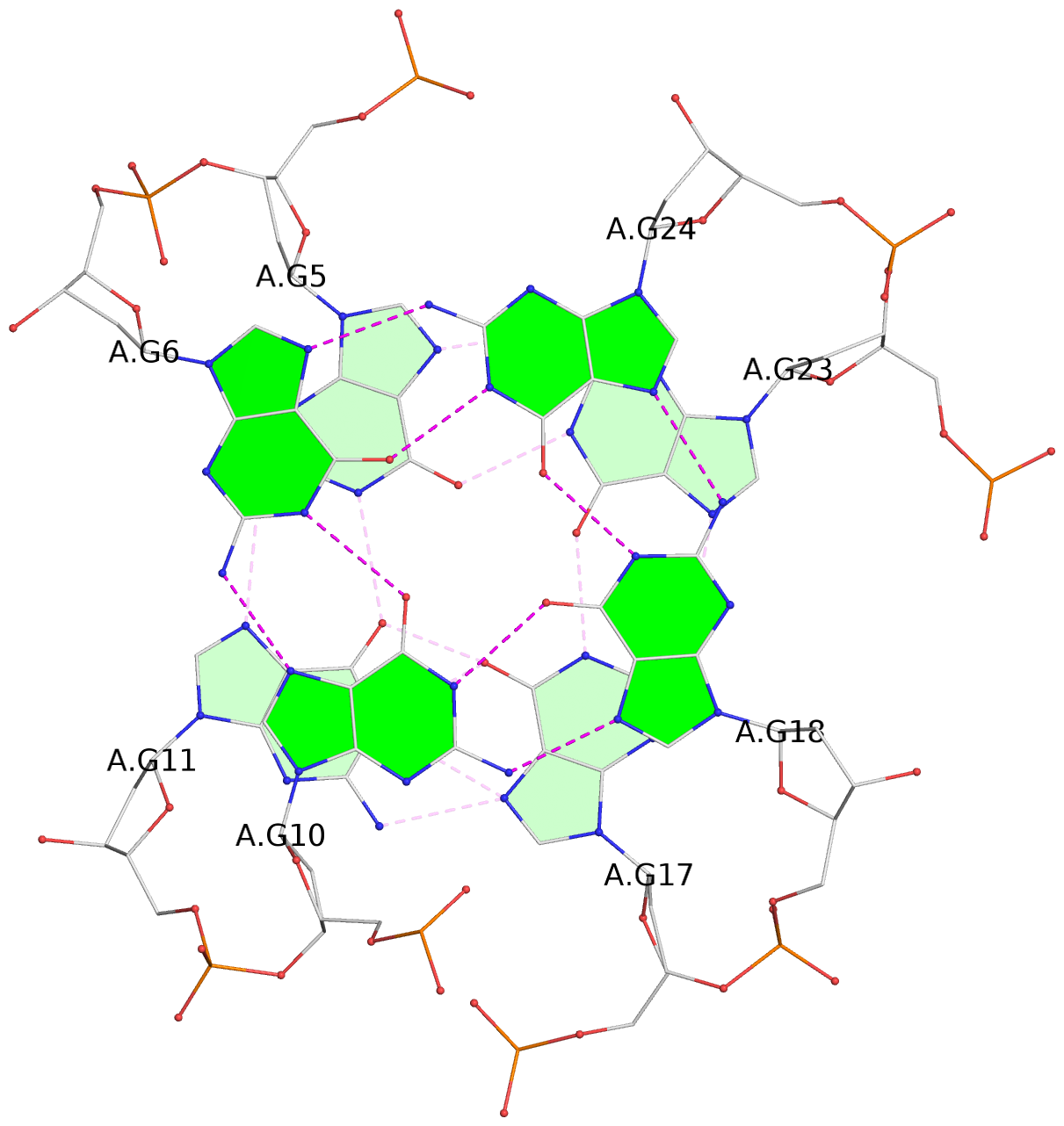
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-Lw-Ln-P), hybrid-2(3+1), UDUU
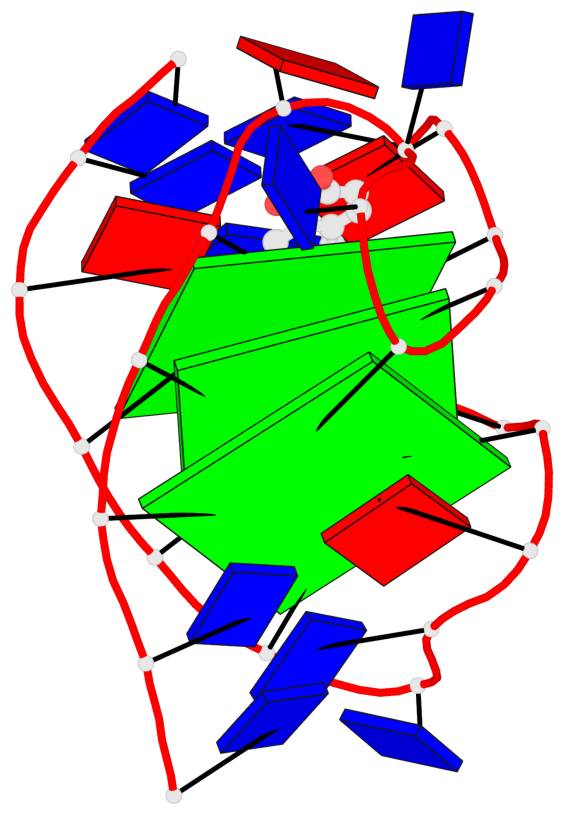
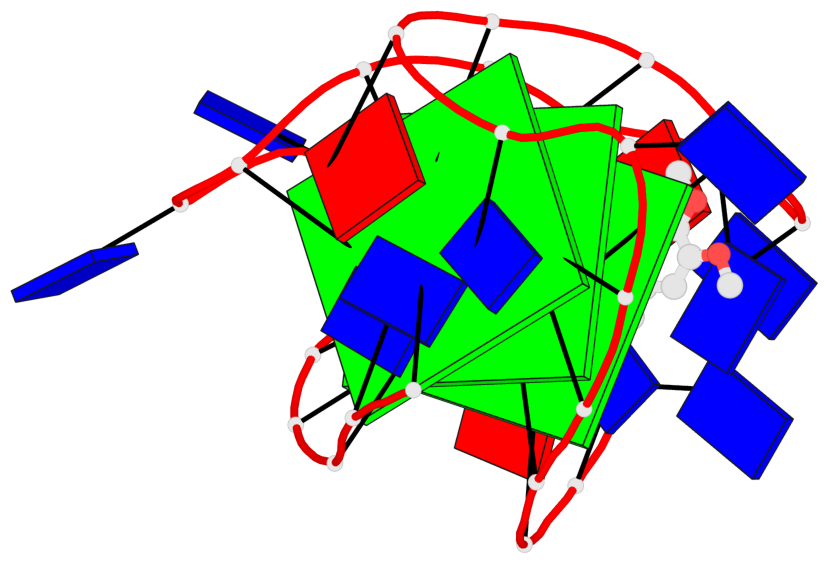
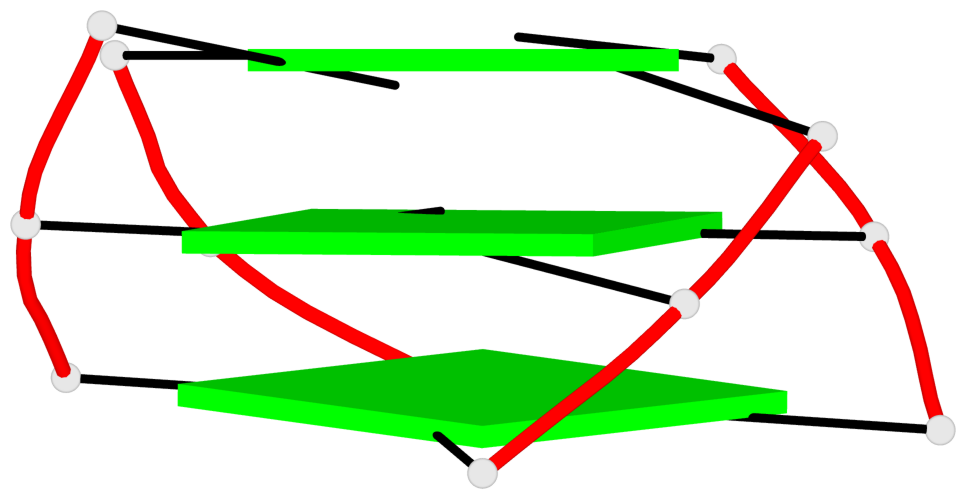
Base-block schematics in six views
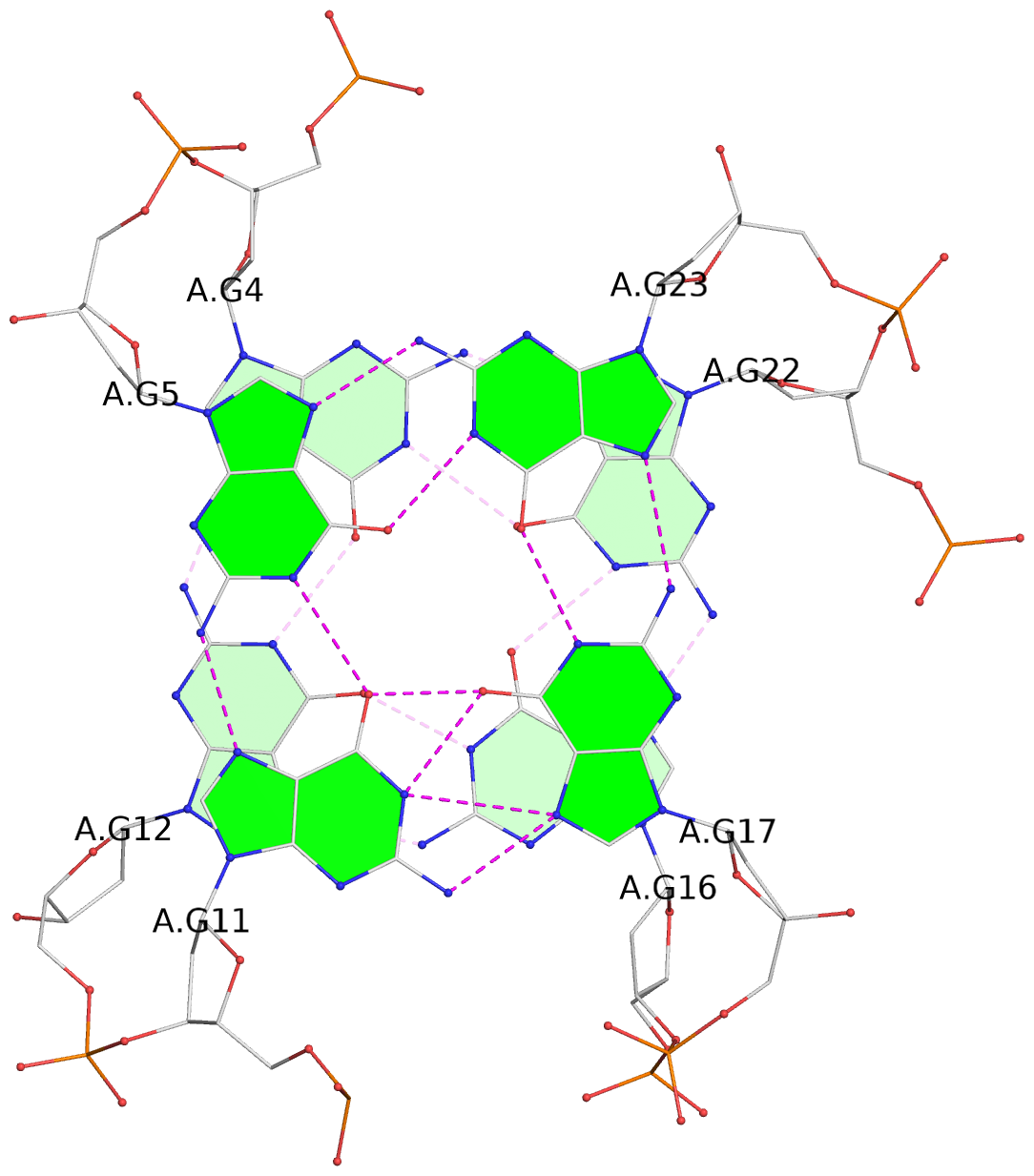
List of 3 G-tetrads
1 glyco-bond=s-ss sugar=-..- groove=wn-- planarity=0.262 type=other nts=4 GGGG A.DG4,A.DG12,A.DG16,A.DG22 2 glyco-bond=-s-- sugar=.-.. groove=wn-- planarity=0.153 type=planar nts=4 GGGG A.DG5,A.DG11,A.DG17,A.DG23 3 glyco-bond=-s-- sugar=--.- groove=wn-- planarity=0.129 type=planar nts=4 GGGG A.DG6,A.DG10,A.DG18,A.DG24
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.