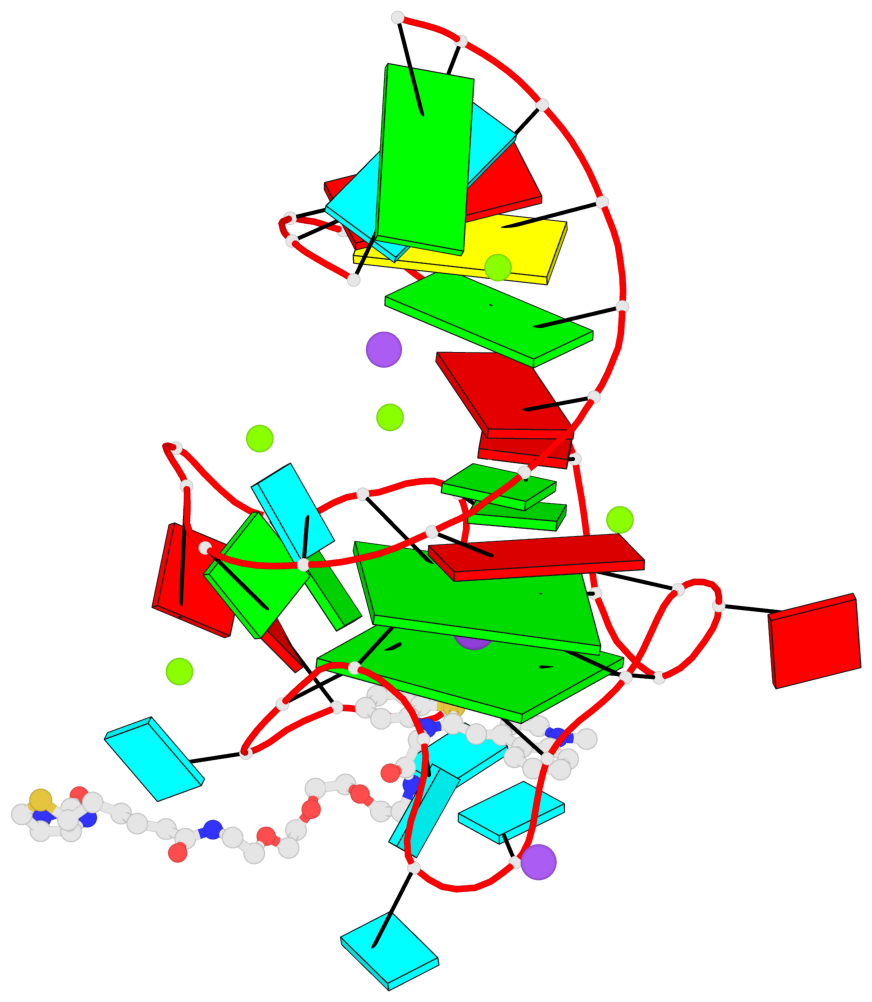
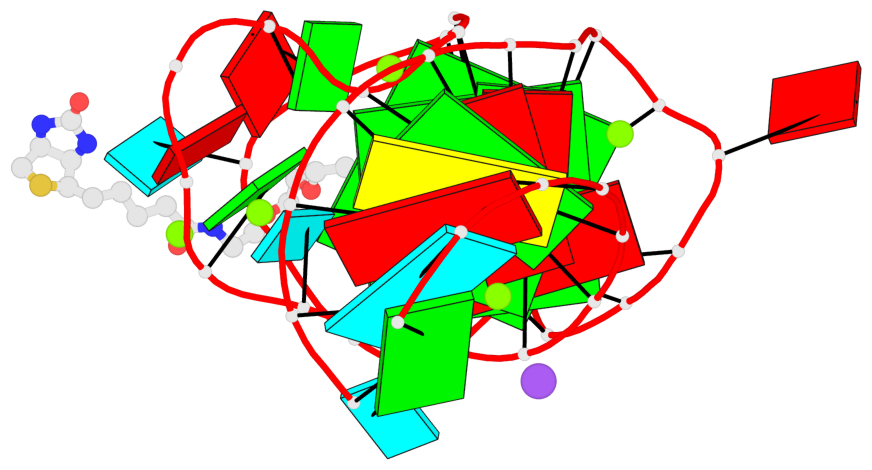
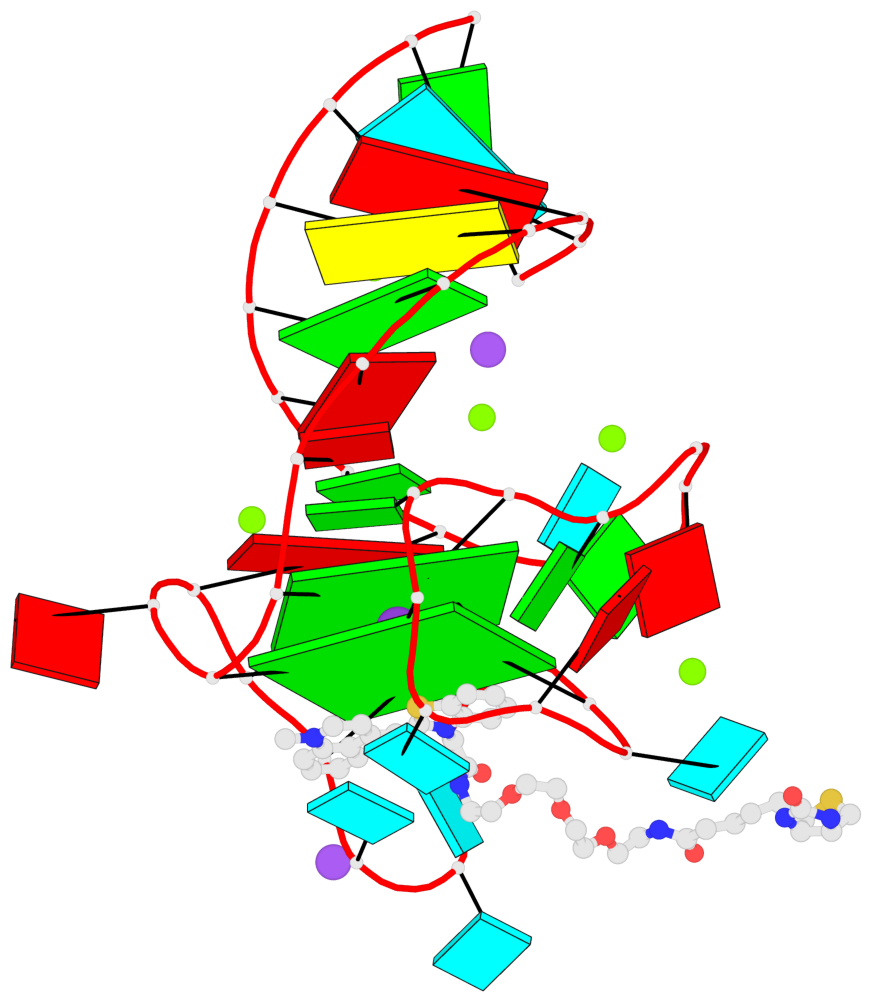
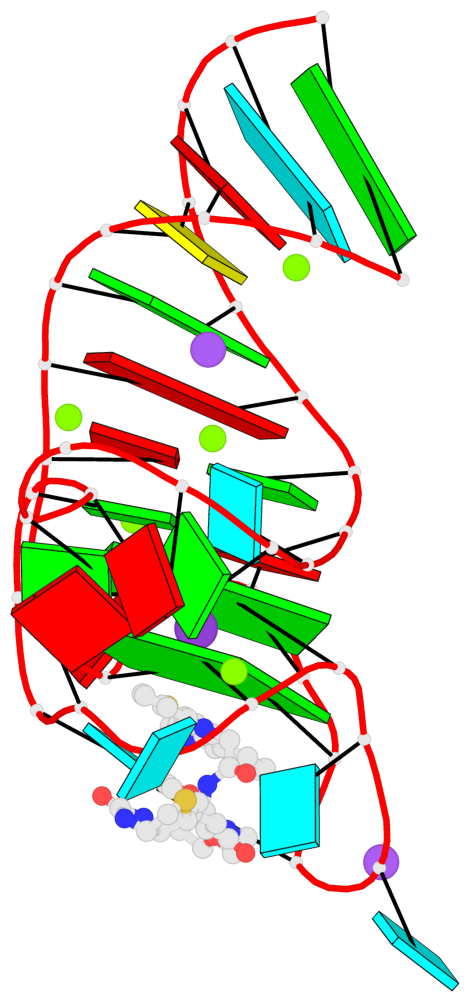
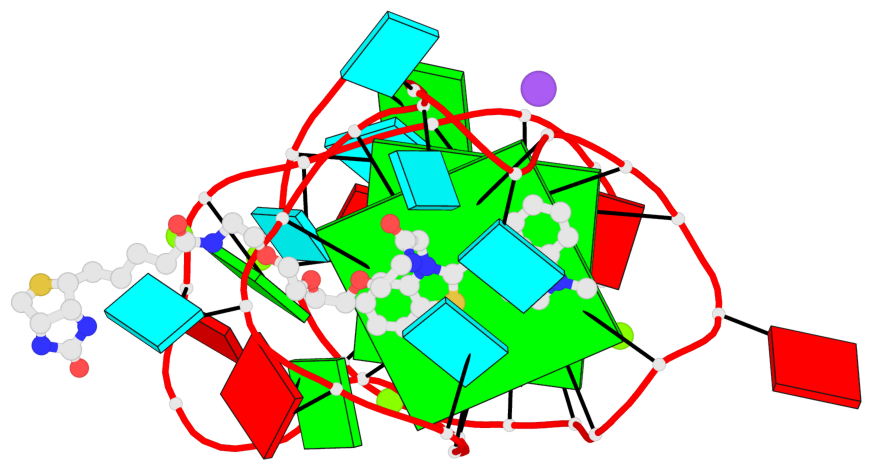
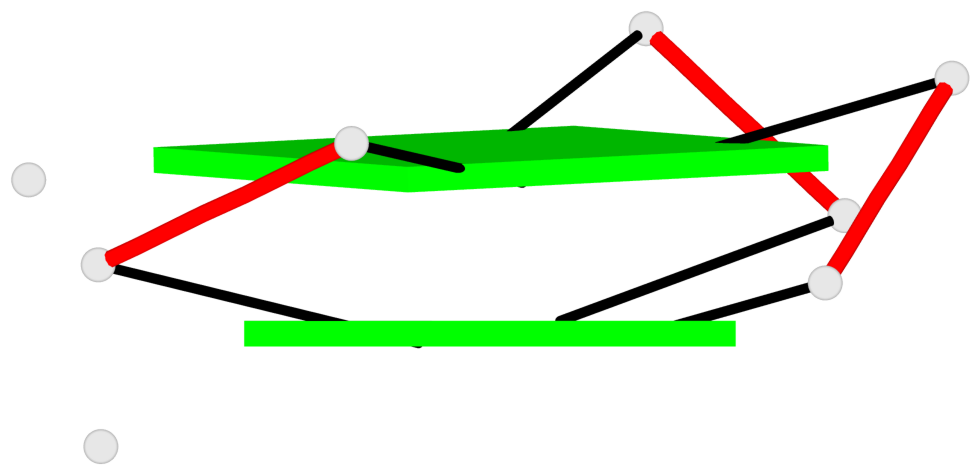
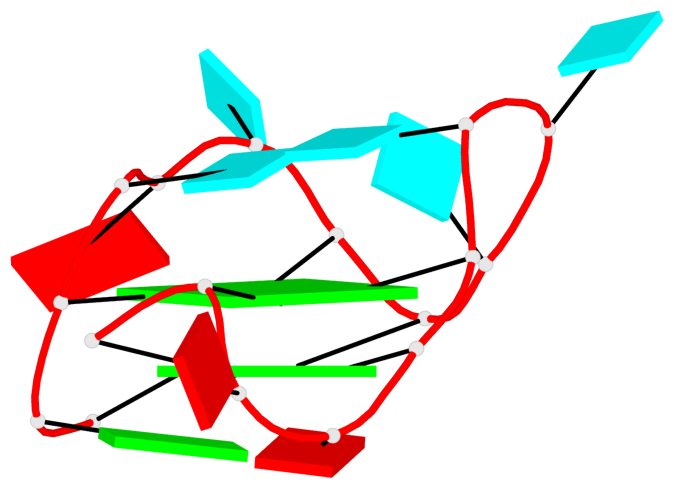
Detailed DSSR results for the G-quadruplex: PDB entry 6e8u
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6e8u
- Class
- RNA
- Method
- X-ray (1.55 Å)
- Summary
- Structure of the mango-iii (a10u) aptamer bound to to1-biotin
- Reference
- Trachman 3rd RJ, Autour A, Jeng SCY, Abdolahzadeh A, Andreoni A, Cojocaru R, Garipov R, Dolgosheina EV, Knutson JR, Ryckelynck M, Unrau PJ, Ferre-D'Amare AR (2019): "Structure and functional reselection of the Mango-III fluorogenic RNA aptamer." Nat. Chem. Biol., 15, 472-479. doi: 10.1038/s41589-019-0267-9.
- Abstract
- Several turn-on RNA aptamers that activate small-molecule fluorophores have been selected in vitro. Among these, the ~30 nucleotide Mango-III is notable because it binds the thiazole orange derivative TO1-Biotin with high affinity and fluoresces brightly (quantum yield 0.55). Uniquely among related aptamers, Mango-III exhibits biphasic thermal melting, characteristic of molecules with tertiary structure. We report crystal structures of TO1-Biotin complexes of Mango-III, a structure-guided mutant Mango-III(A10U), and a functionally reselected mutant iMango-III. The structures reveal a globular architecture arising from an unprecedented pseudoknot-like connectivity between a G-quadruplex and an embedded non-canonical duplex. The fluorophore is restrained into a planar conformation by the G-quadruplex, a lone, long-range trans Watson-Crick pair (whose A10U mutation increases quantum yield to 0.66), and a pyrimidine perpendicular to the nucleobase planes of those motifs. The improved iMango-III and Mango-III(A10U) fluoresce ~50% brighter than enhanced green fluorescent protein, making them suitable tags for live cell RNA visualization.
- G4 notes
- 2 G-tetrads, 1 G4 helix, 1 G4 stem, 2(-P-P-Lw), hybrid-2R(3+1), UUUD
Base-block schematics in six views
List of 2 G-tetrads
1 glyco-bond=---s sugar=-33- groove=--wn planarity=0.321 type=other nts=4 GGGG B.G8,B.G12,B.G17,B.G24 2 glyco-bond=---s sugar=-3-3 groove=--wn planarity=0.260 type=other nts=4 GGGG B.G9,B.G13,B.G18,B.G22
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.