Detailed DSSR results for the G-quadruplex: PDB entry 6eo6
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6eo6
- Class
- hydrolase-DNA
- Method
- X-ray (1.69 Å)
- Summary
- X-ray structure of the complex between human alpha-thrombin and modified 15-mer DNA aptamer containing 5-(3-(2-(1h-indol-3-yl)acetamide-n-yl)-1-propen-1-yl)-2'-deoxyuridine residue
- Reference
- Dolot R, Lam CH, Sierant M, Zhao Q, Liu FW, Nawrot B, Egli M, Yang X (2018): "Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity." Nucleic Acids Res., 46, 4819-4830. doi: 10.1093/nar/gky268.
- Abstract
- Thrombin-binding aptamer (TBA) is a DNA 15-mer of sequence 5'-GGT TGG TGT GGT TGG-3' that folds into a G-quadruplex structure linked by two T-T loops located on one side and a T-G-T loop on the other. These loops are critical for post-SELEX modification to improve TBA target affinity. With this goal in mind we synthesized a T analog, 5-(indolyl-3-acetyl-3-amino-1-propenyl)-2'-deoxyuridine (W) to substitute one T or a pair of Ts. Subsequently, the affinity for each analog was determined by biolayer interferometry. An aptamer with W at position 4 exhibited about 3-fold increased binding affinity, and replacing both T4 and T12 with W afforded an almost 10-fold enhancement compared to native TBA. To better understand the role of the substituent's aromatic moiety, an aptamer with 5-(methyl-3-acetyl-3-amino-1-propenyl)-2'-deoxyuridine (K; W without the indole moiety) in place of T4 was also synthesized. This K4 aptamer was found to improve affinity 7-fold relative to native TBA. Crystal structures of aptamers with T4 replaced by either W or K bound to thrombin provide insight into the origins of the increased affinities. Our work demonstrates that facile chemical modification of a simple DNA aptamer can be used to significantly improve its binding affinity for a well-established pharmacological target protein.
- G4 notes
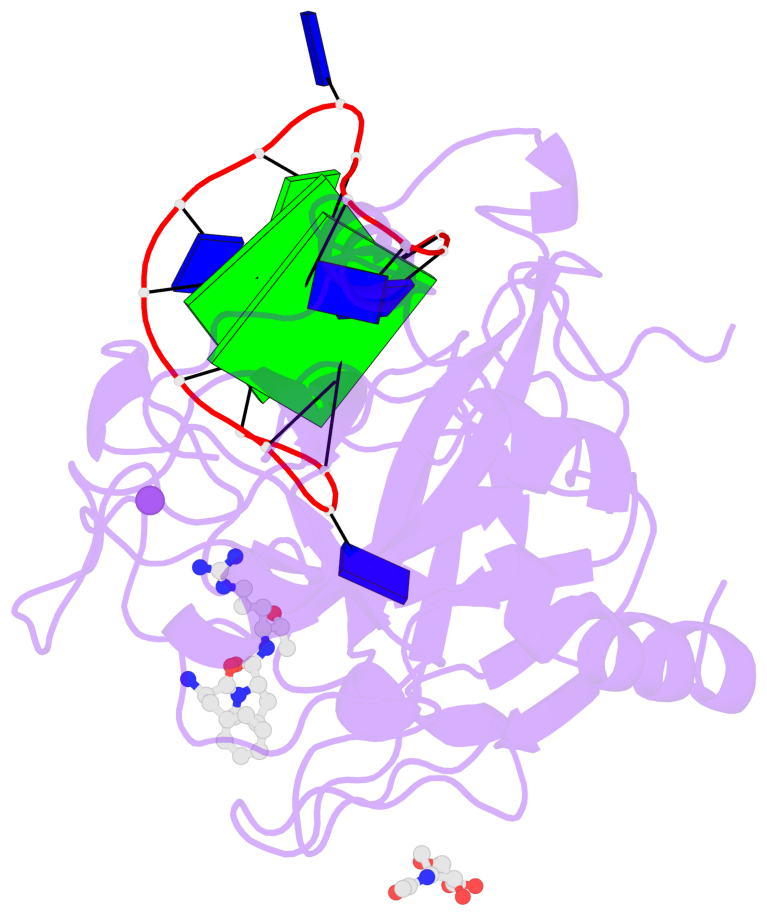
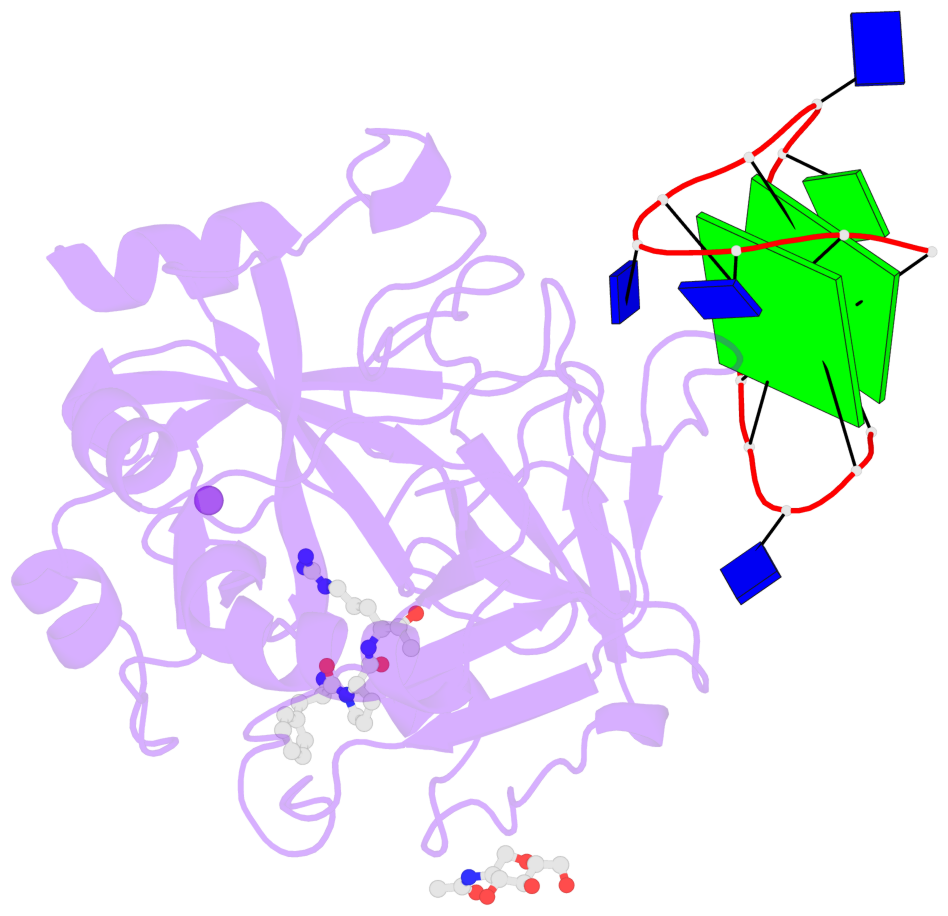
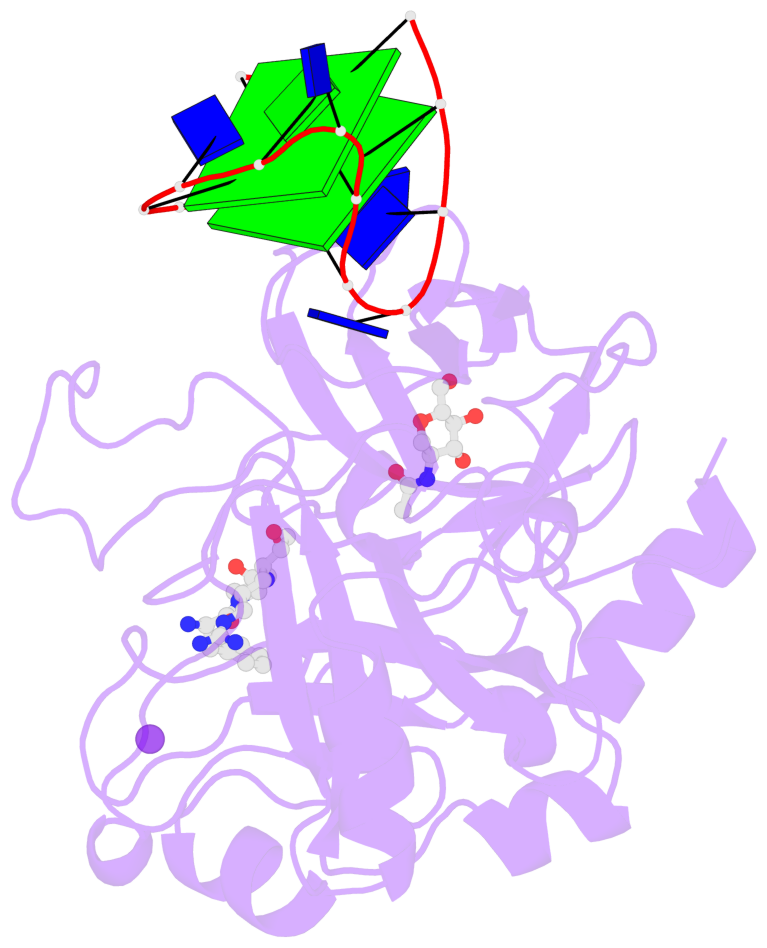
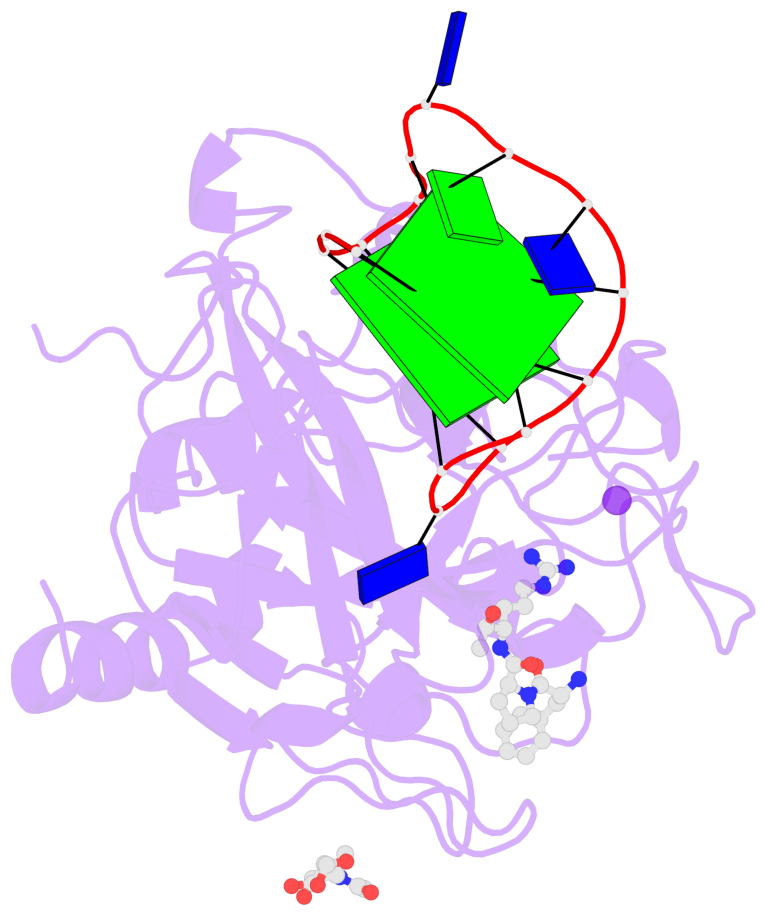
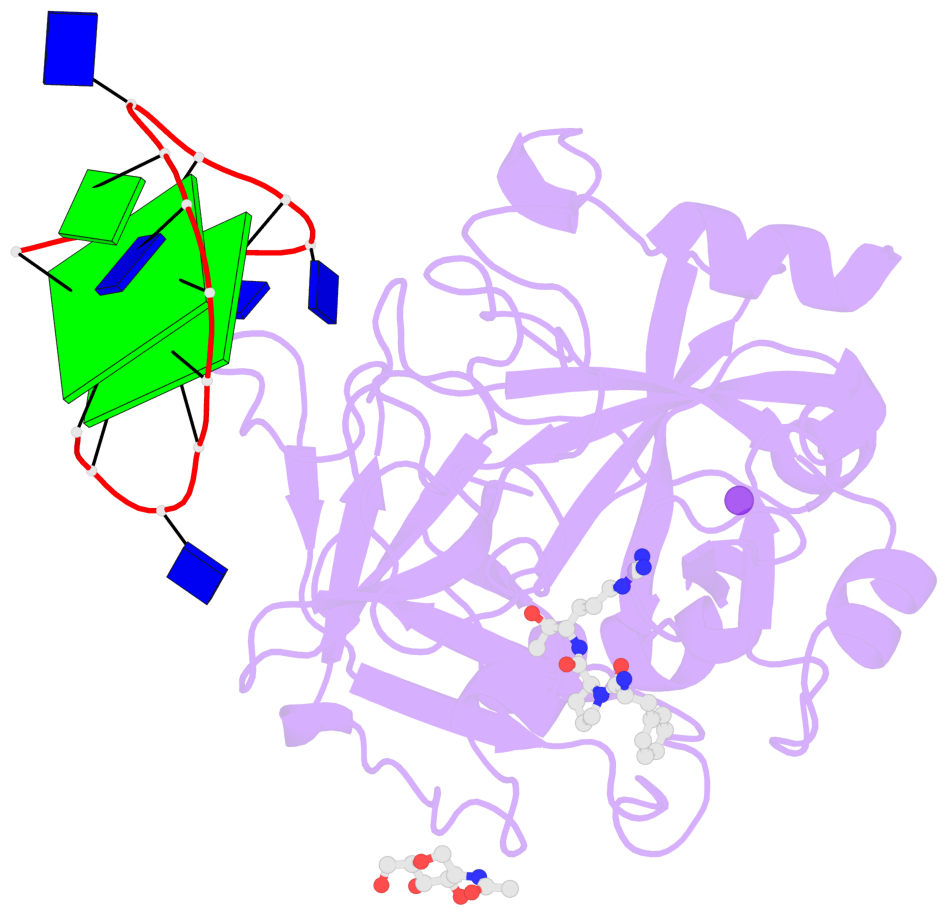
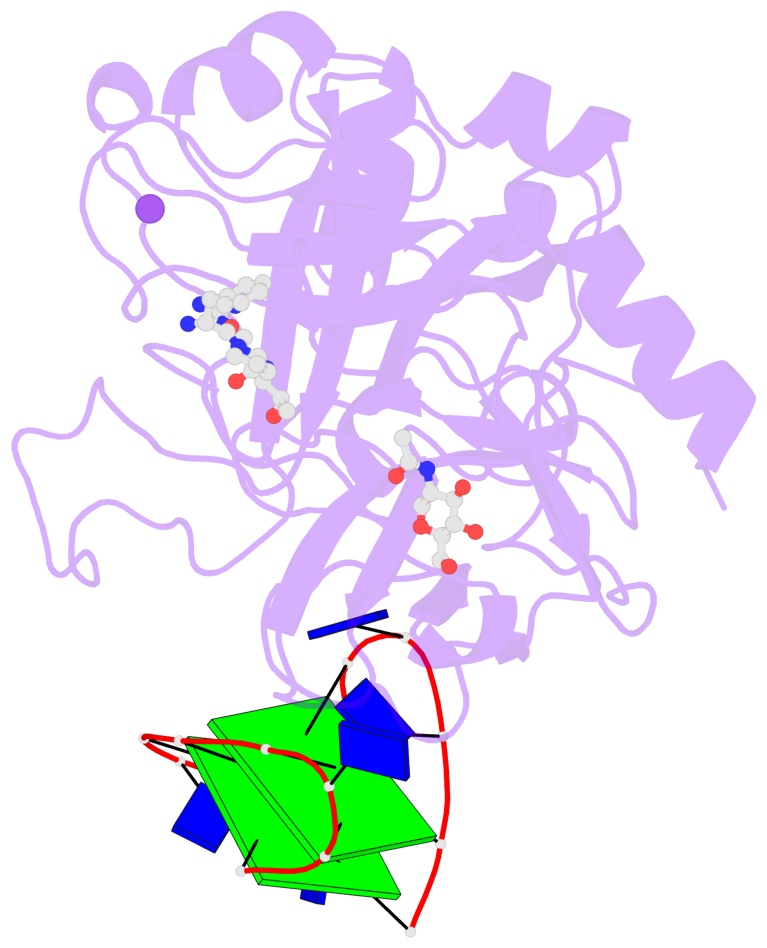
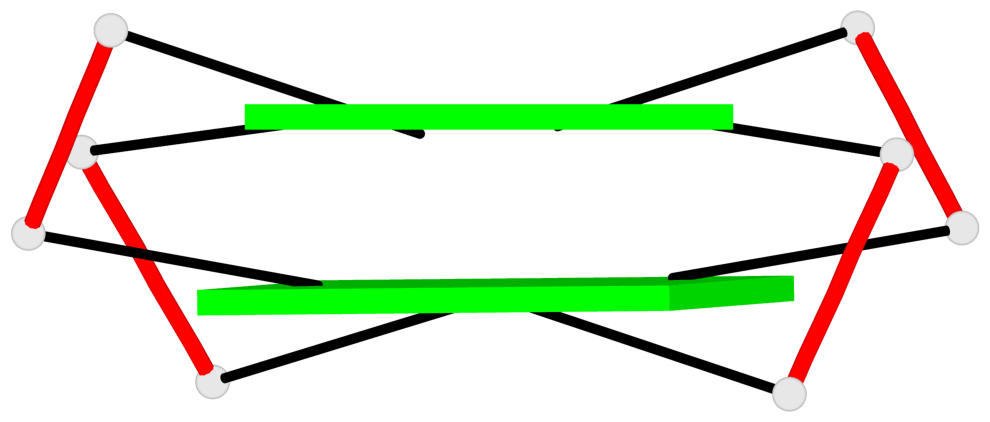
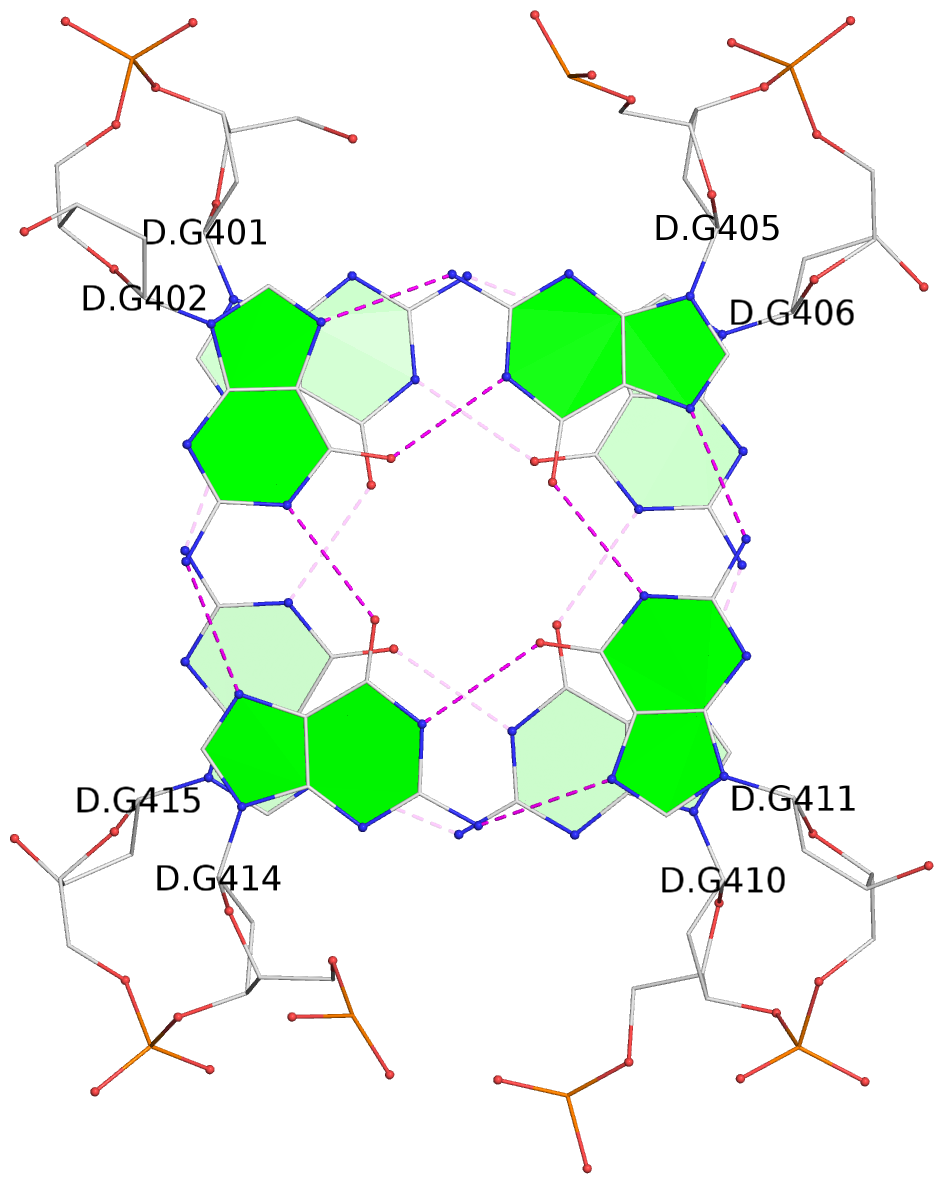
- 2 G-tetrads, 1 G4 helix, 1 G4 stem, 2(+Ln+Lw+Ln), chair(2+2), UDUD
Base-block schematics in six views
List of 2 G-tetrads
1 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.499 type=saddle nts=4 GGGG D.DG401,D.DG415,D.DG410,D.DG406 2 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.562 type=other nts=4 GGGG D.DG402,D.DG414,D.DG411,D.DG405
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.