Detailed DSSR results for the G-quadruplex: PDB entry 6erl
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6erl
- Class
- DNA
- Method
- NMR
- Summary
- Quadruplex with flipped tetrad formed by the c-myc promoter sequence
- Reference
- Karg B, Weisz K (2018): "Loop Length Affects Syn-Anti Conformational Rearrangements in Parallel G-Quadruplexes." Chemistry. doi: 10.1002/chem.201801851.
- Abstract
- A G-quadruplex forming sequence from the MYC promoter region was modified with syn-favoring 8-bromo-2'-deoxyguanosine residues. Depending on the number and position of modifications in the intramolecular parallel G-quadruplex, substitutions with the bromoguanosine analogue at the 5'-tetrad induce conformational rearrangements with concerted all-anti to all-syn transitions for all residues of the modified G-quartet. No unfavorable steric interactions of the C8-substituents in the medium grooves are apparent in the high-resolution structure as determined for a tetrasubstituted MYC quadruplex that exclusively forms the all-syn isomer. In contrast, considerable steric clashes with 5'-phosphate oxygen atoms for those analogues that follow a less flexible 1-nucleotide loop in the native all-anti conformation seem to constitute the major driving force for the tetrad inversion and allow for the rational design of appropriately substituted sequences. Correlations found between the population of species subjected to a tetrad flip and melting temperatures indicate that more effective conformational transitions are compromised by lower thermal stabilities of the modified parallel quadruplexes.
- G4 notes
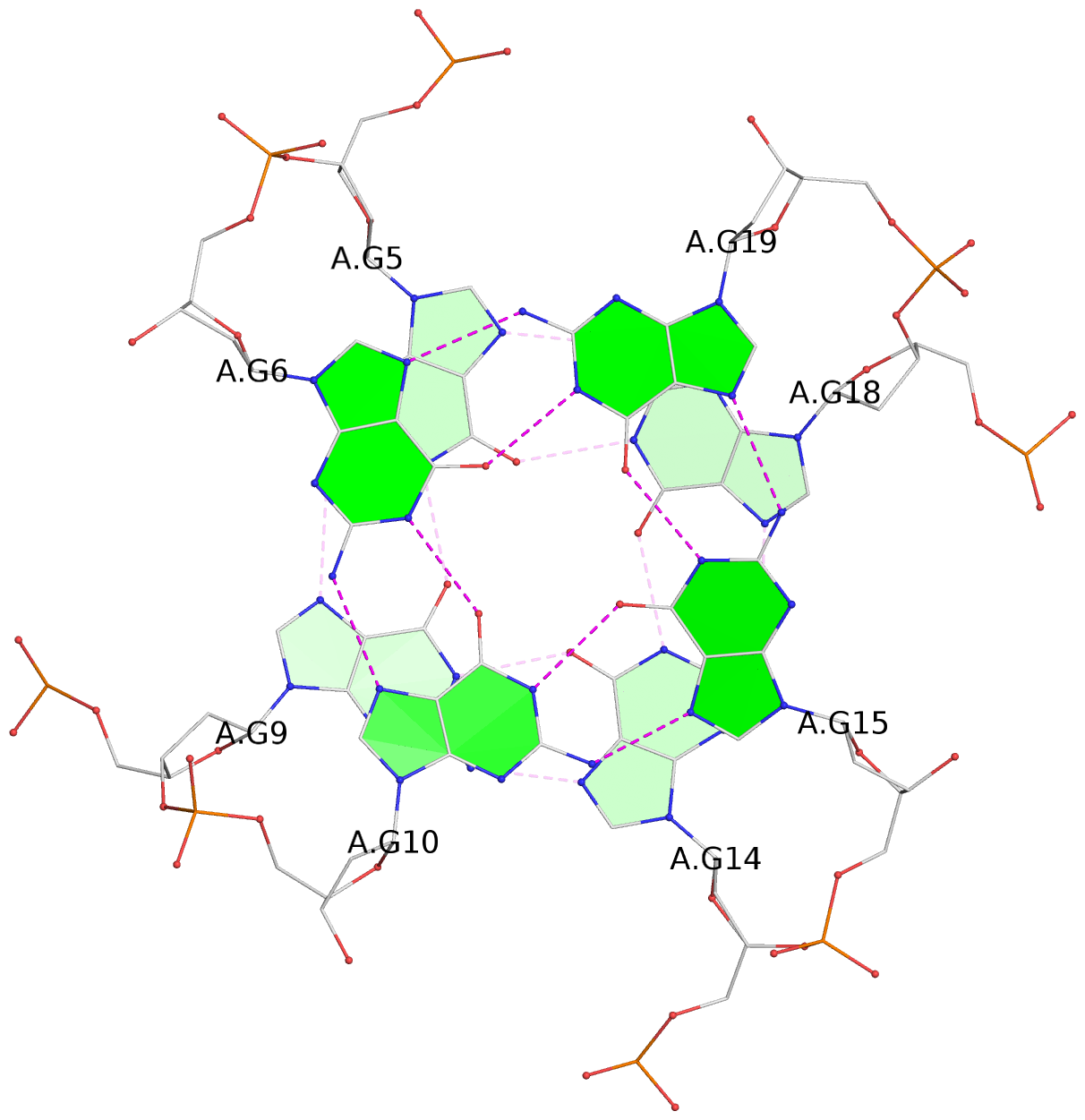
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
Base-block schematics in six views
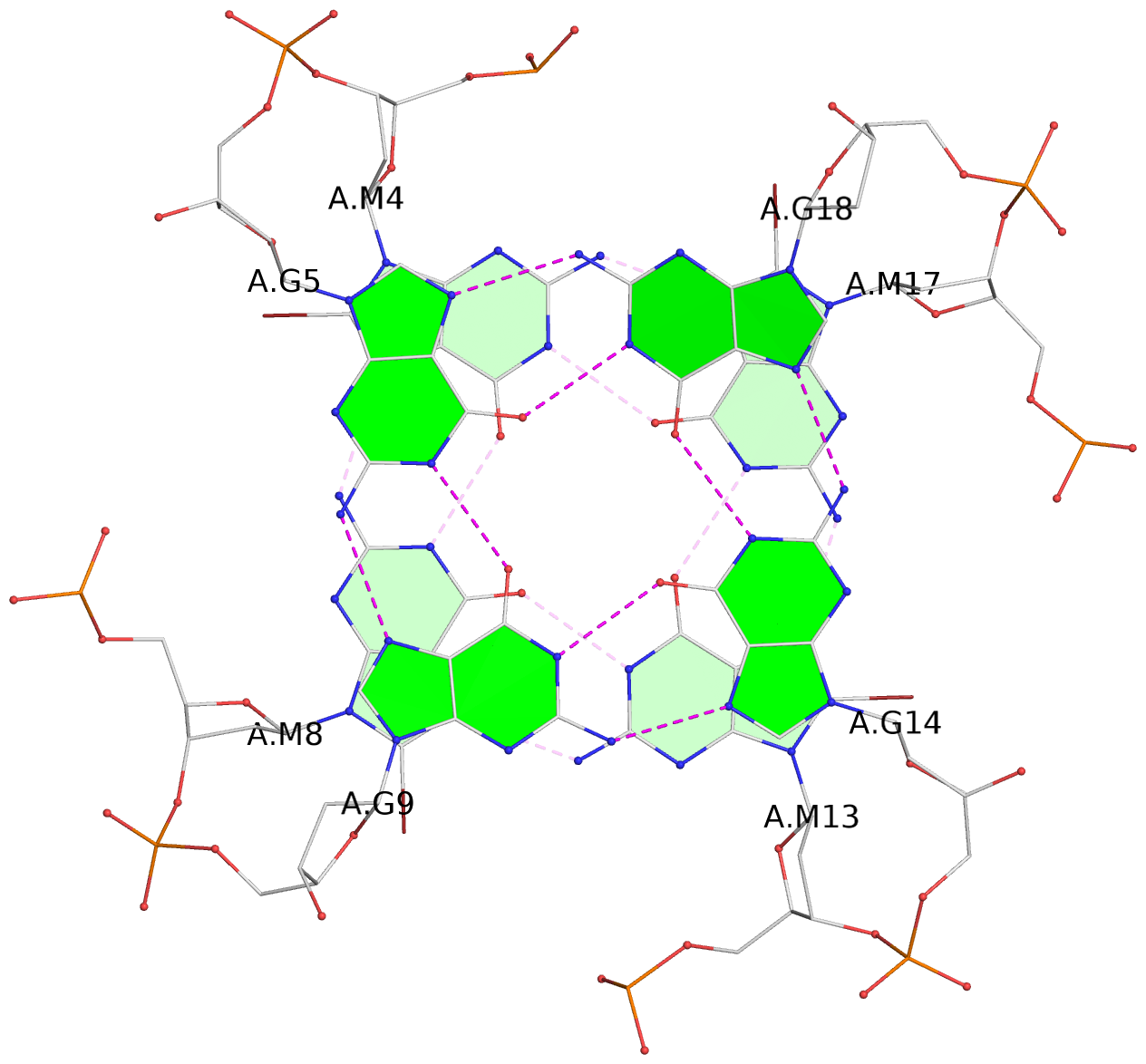
List of 3 G-tetrads
1 glyco-bond=ssss sugar=---- groove=---- planarity=0.326 type=other nts=4 gggg A.BGM4,A.BGM8,A.BGM13,A.BGM17 2 glyco-bond=---- sugar=---- groove=---- planarity=0.465 type=other nts=4 GGGG A.DG5,A.DG9,A.DG14,A.DG18 3 glyco-bond=---- sugar=---- groove=---- planarity=0.419 type=bowl nts=4 GGGG A.DG6,A.DG10,A.DG15,A.DG19
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.