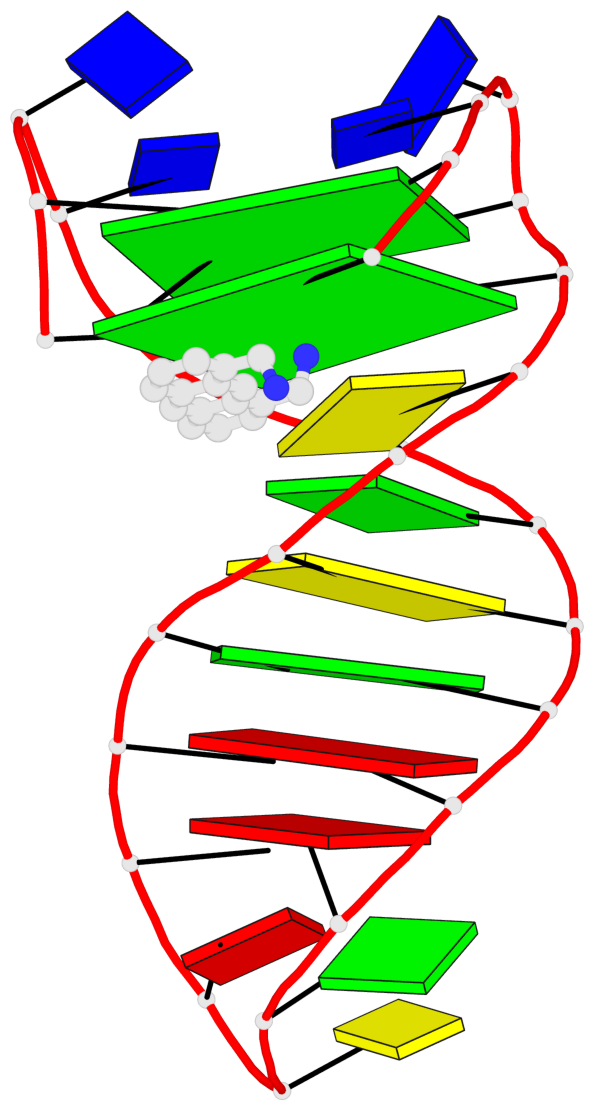
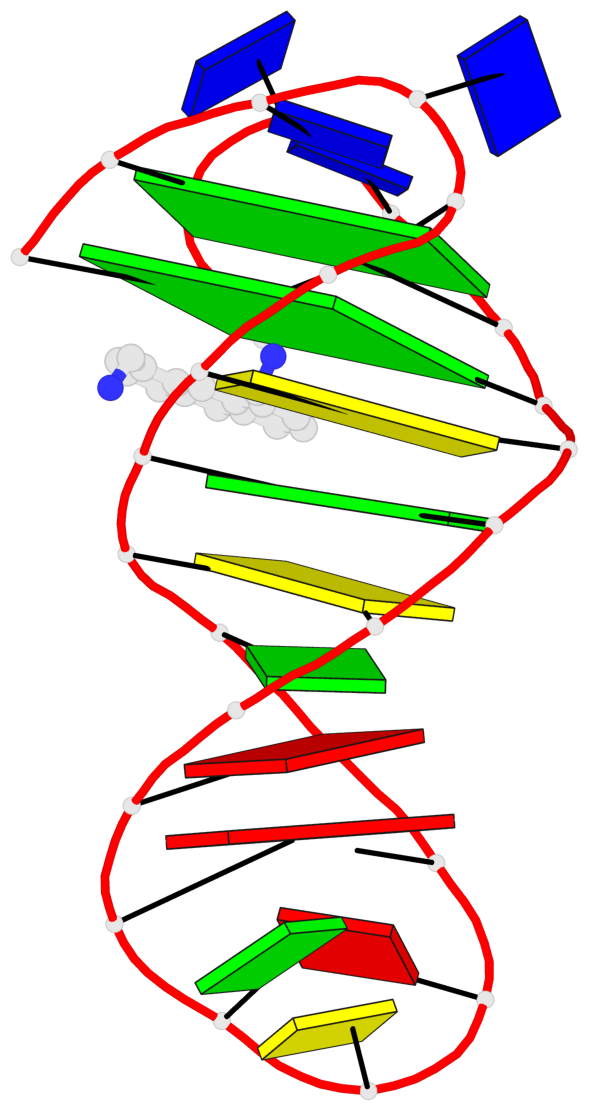
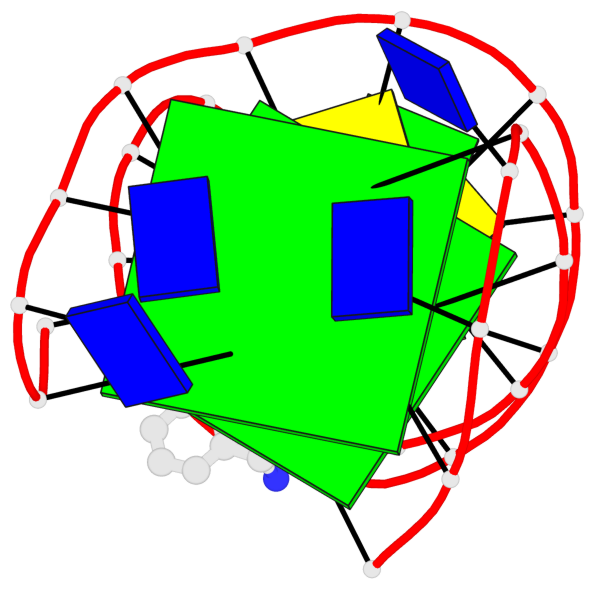
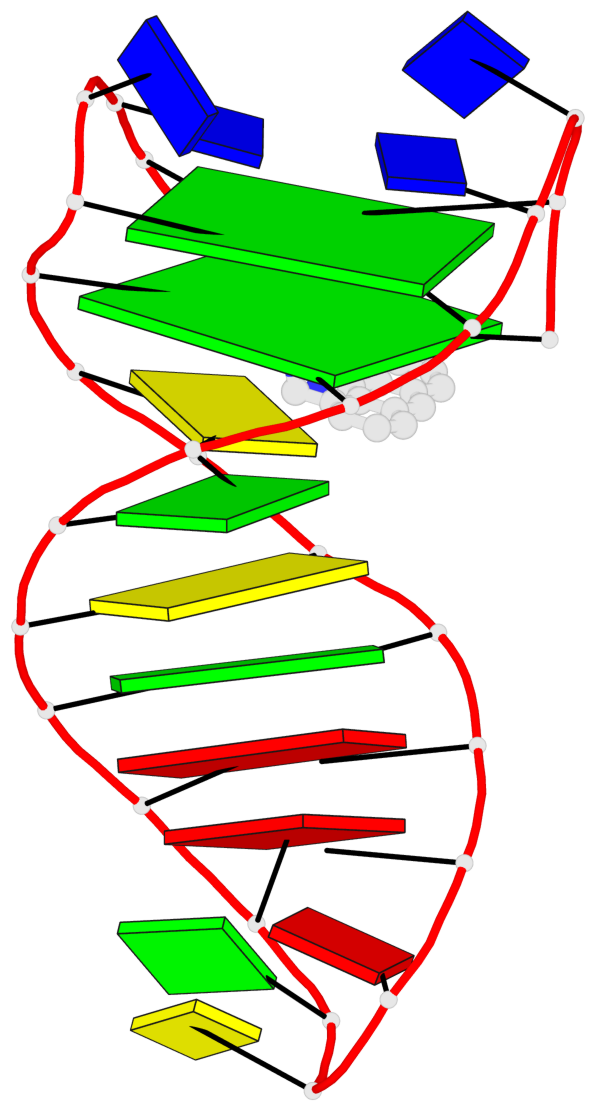
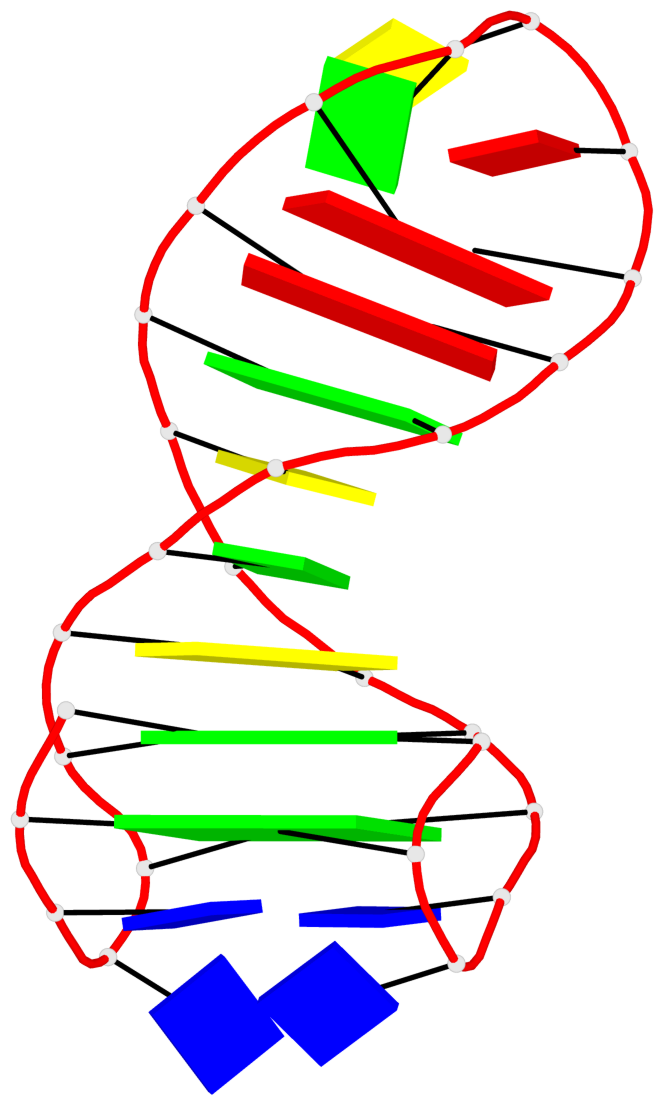
Detailed DSSR results for the G-quadruplex: PDB entry 6fc9
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6fc9
- Class
- DNA
- Method
- NMR
- Summary
- The 1,8-bis(aminomethyl)anthracene and quadruplex-duplex junction complex
- Reference
- Diaz-Casado L, Serrano-Chacon I, Montalvillo-Jimenez L, Corzana F, Bastida A, Santana AG, Gonzalez C, Asensio JL (2020): "De Novo Design of Selective Quadruplex-Duplex Junction Ligands and Structural Characterisation of Their Binding Mode: Targeting the G4 Hot-Spot." Chemistry. doi: 10.1002/chem.202005026.
- Abstract
- Targeting the interface between DNA quadruplex and duplex regions by small molecules holds significant promise in both therapeutics and nanotechnology. Herein, a new pharmacophore is reported, which selectively binds with high affinity to quadruplex-duplex junctions, while presenting a poorer affinity for G-quadruplex or duplex DNA alone. Ligands complying with the reported pharmacophore exhibit a significant affinity and selectivity for quadruplex-duplex junctions, including the one observed in the HIV-1 LTR-III sequence. The structure of the complex between a quadruplex-duplex junction with a ligand of this family has been determined by NMR methods. According to these data, the remarkable selectivity of this structural motif for quadruplex-duplex junctions is achieved through an unprecedented interaction mode so far unexploited in medicinal and biological chemistry: the insertion of a benzylic ammonium moiety into the centre of the partially exposed G-tetrad at the interface with the duplex. Further decoration of the described scaffolds with additional fragments opens up the road to the development of selective ligands for G-quadruplex-forming regions of the genome.
- G4 notes
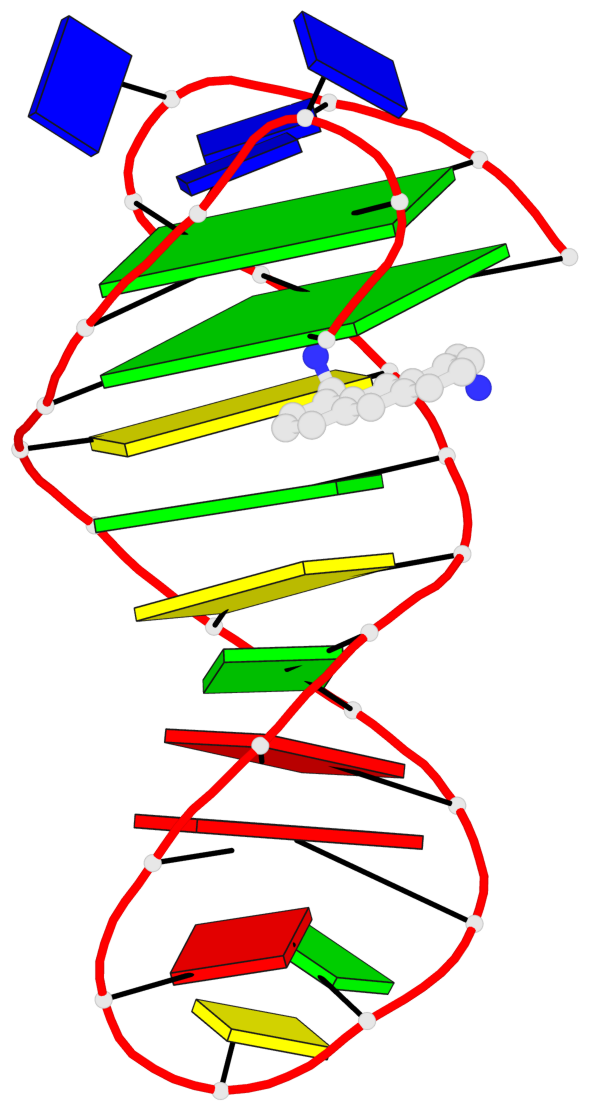
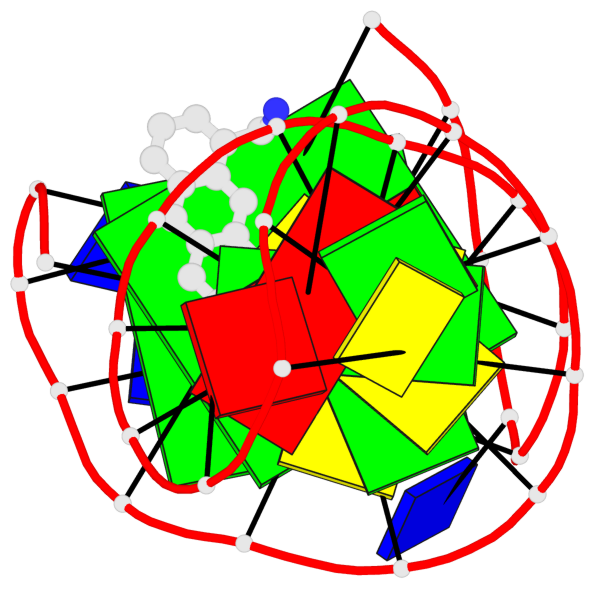
- 2 G-tetrads, 1 G4 helix, 1 G4 stem, 2(+Ln+Lw+Ln), chair(2+2), UDUD
Base-block schematics in six views
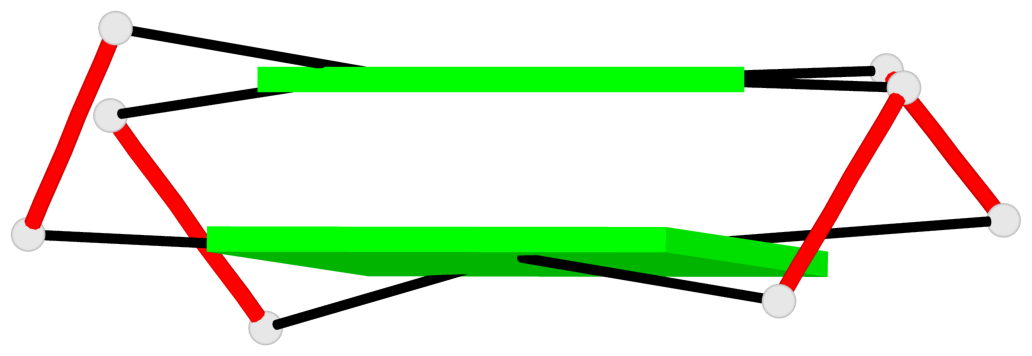
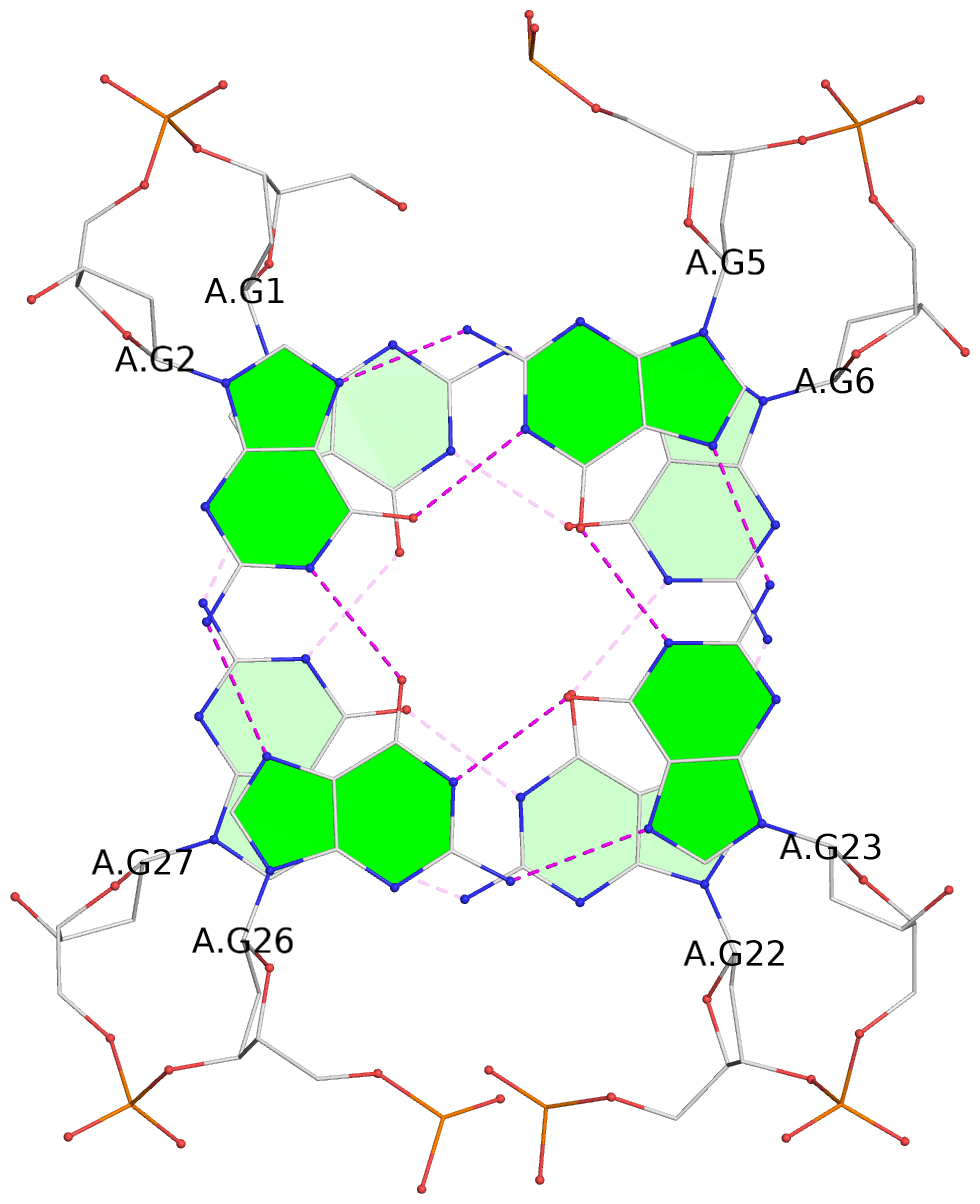
List of 2 G-tetrads
1 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.172 type=other nts=4 GGGG A.DG1,A.DG27,A.DG22,A.DG6 2 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.213 type=other nts=4 GGGG A.DG2,A.DG26,A.DG23,A.DG5
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.