Detailed DSSR results for the G-quadruplex: PDB entry 6ffr
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6ffr
- Class
- DNA-RNA hybrid
- Method
- NMR
- Summary
- DNA-RNA hybrid quadruplex with flipped tetrad
- Reference
- Haase L, Dickerhoff J, Weisz K (2018): "DNA-RNA Hybrid Quadruplexes Reveal Interactions that Favor RNA Parallel Topologies." Chemistry, 24, 15365-15371. doi: 10.1002/chem.201803367.
- Abstract
- A DNA G-quadruplex adopting a (3+1)-hybrid structure was substituted at its 5'-tetrad by riboguanosine (rG) analogs. Incorporation of anti-favoring rG at appropriate syn-positions of the 5'-outer tetrad induced conformational rearrangements to yield a quadruplex featuring a 5'-tetrad with reversed polarity. A high-resolution structure of a disubstituted quadruplex variant as well as direct NMR experimental evidence reveals a non-conventional C-H⋅⋅⋅O hydrogen bond in a medium groove between the 2'-OH of an rG residue adopting a C2'-endo sugar pucker and H8 of a 3'-neighboring anti-G residue. In contrast, a C3'-endo sugar conformation for another guanine ribonucleotide prevents formation of a corresponding hydrogen bond but relocates its 2'-OH substituent from the quadruplex narrow groove into a medium groove. Both the formation of favorable CHO hydrogen bridges and unfavorable interactions of the 2'-hydroxyl group in a narrow groove will promote RNA folding into a parallel topology featuring all-anti core residues and four grooves of medium size.
- G4 notes
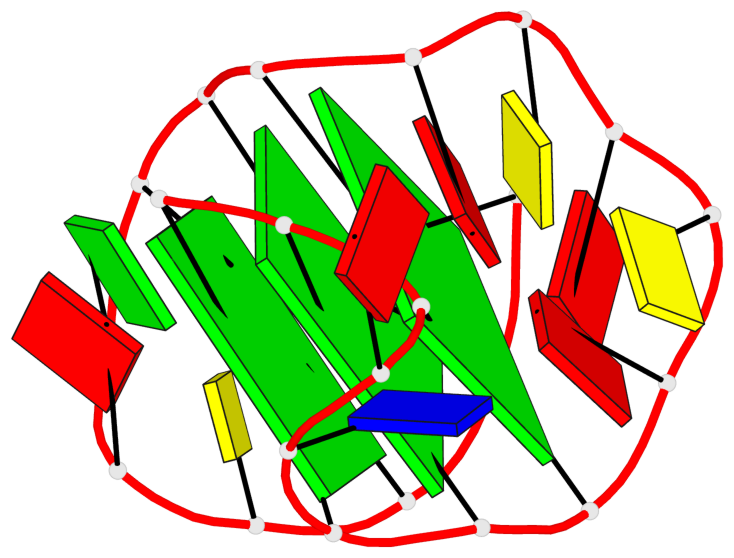
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-PD+Lw), U3D(3+1), UUUD
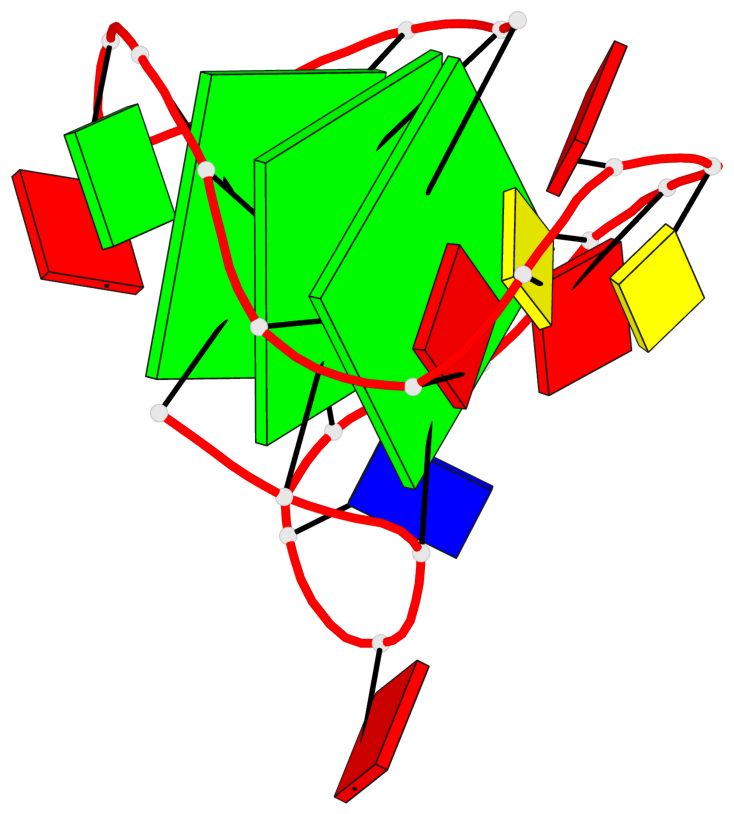
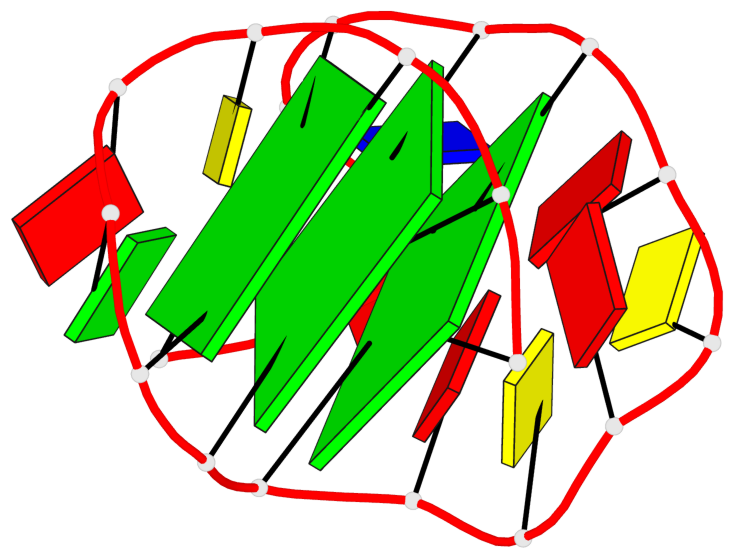
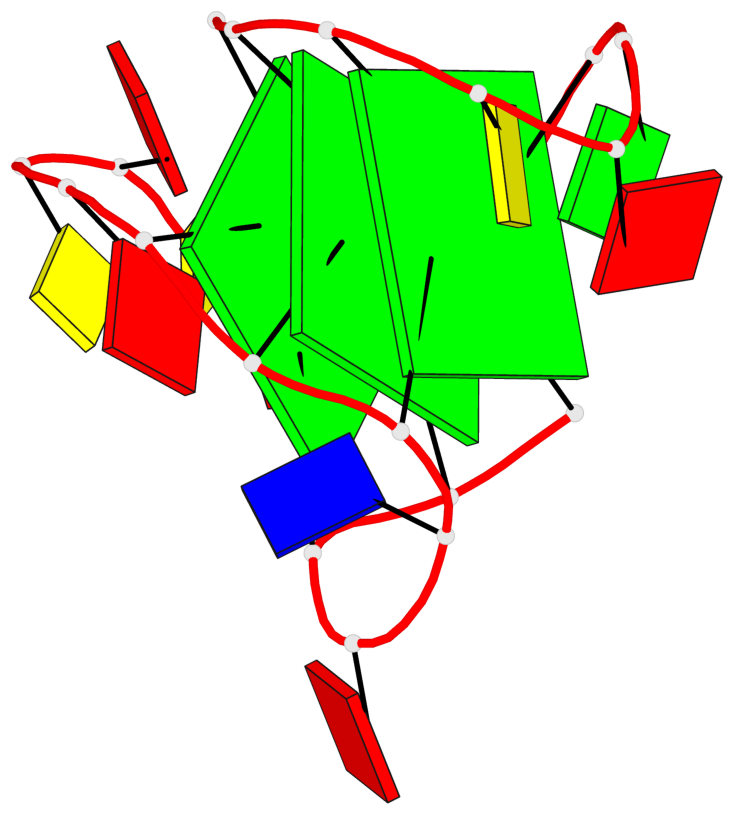
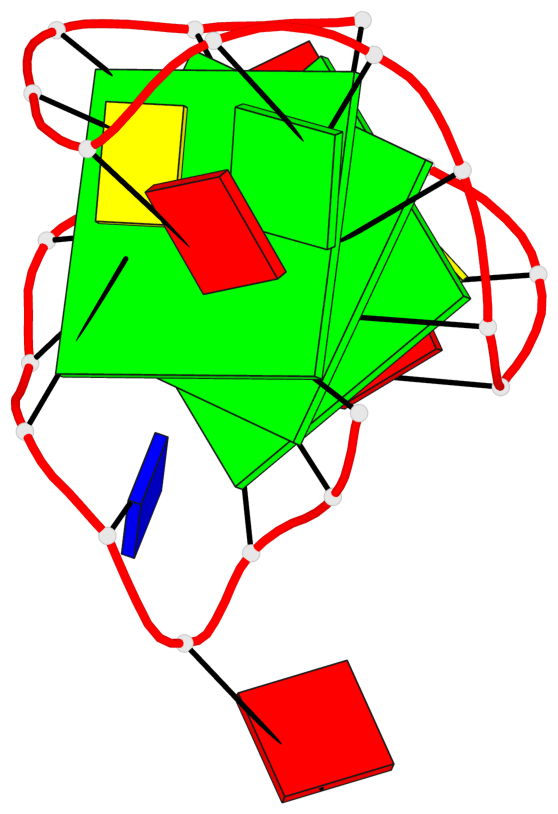
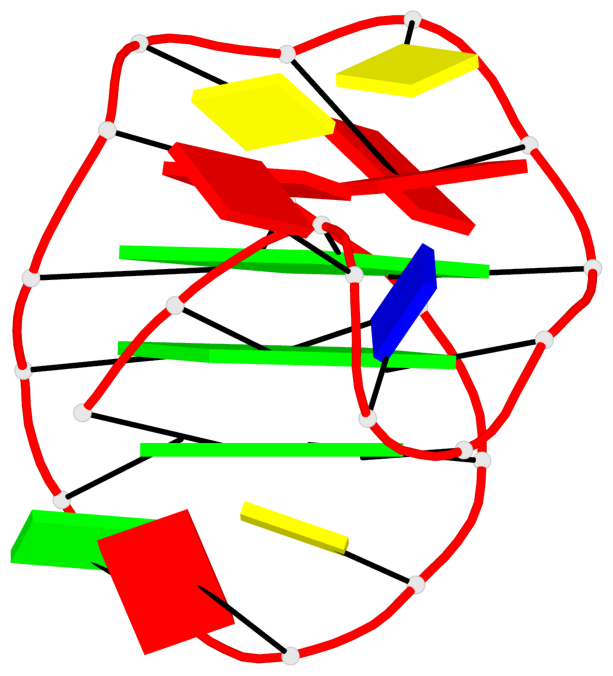
Base-block schematics in six views
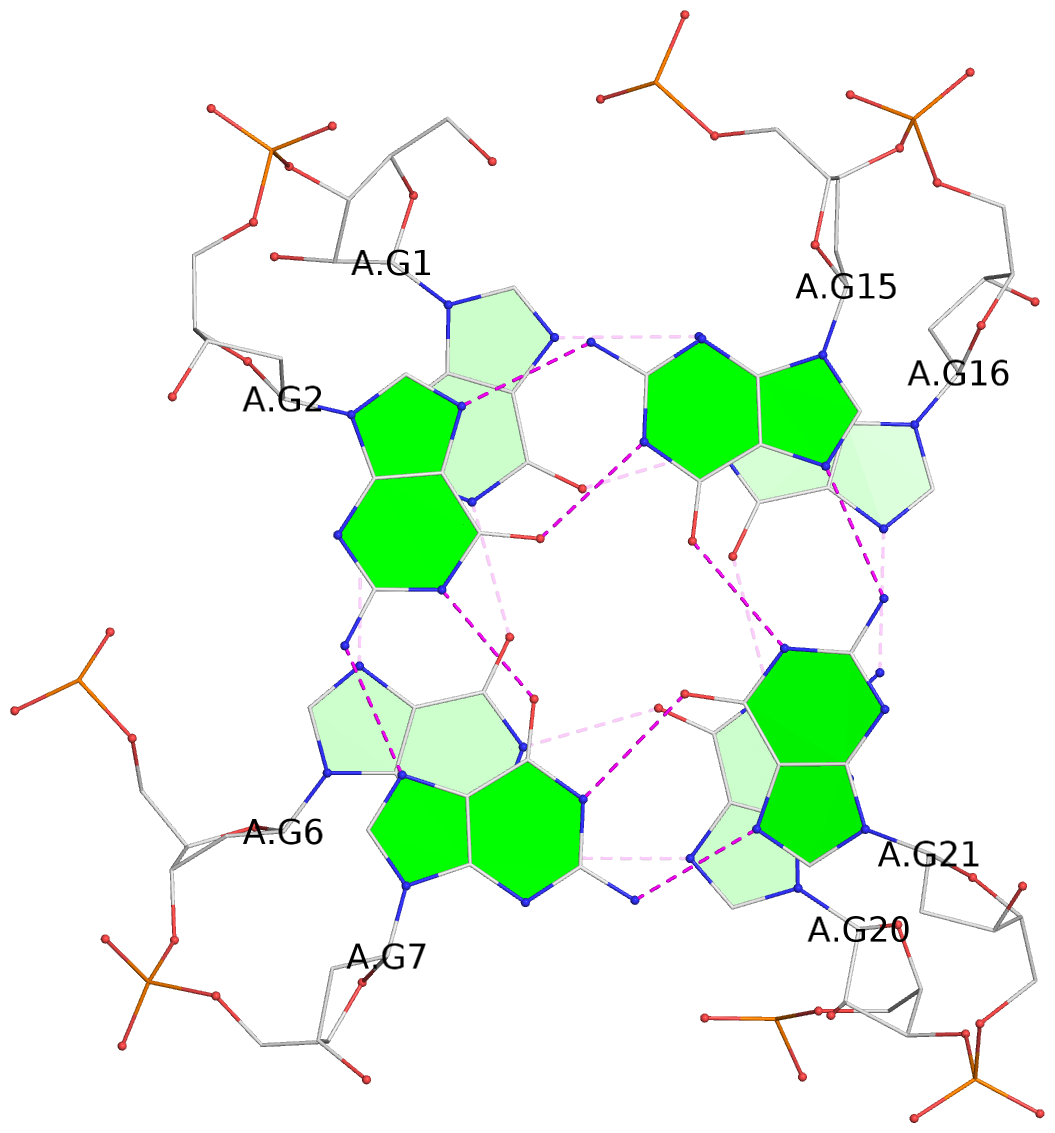
List of 3 G-tetrads
1 glyco-bond=---s sugar=3--- groove=--wn planarity=0.243 type=other nts=4 GGGG A.G1,A.DG6,A.G20,A.DG16 2 glyco-bond=---s sugar=---- groove=--wn planarity=0.253 type=other nts=4 GGGG A.DG2,A.DG7,A.DG21,A.DG15 3 glyco-bond=---s sugar=---- groove=--wn planarity=0.342 type=bowl-2 nts=4 GGGG A.DG3,A.DG8,A.DG22,A.DG14
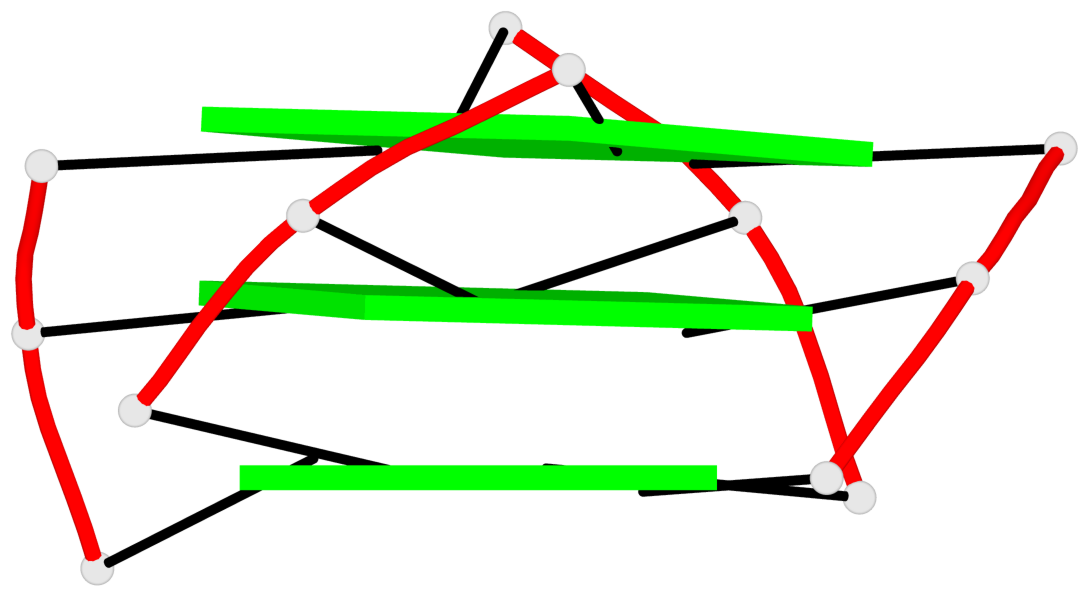
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.