Detailed DSSR results for the G-quadruplex: PDB entry 6fq2
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6fq2
- Class
- DNA
- Method
- X-ray (2.31 Å)
- Summary
- Structure of minimal sequence for left -handed g-quadruplex formation
- Reference
- Bakalar B, Heddi B, Schmitt E, Mechulam Y, Phan AT (2019): "A Minimal Sequence for Left-Handed G-Quadruplex Formation." Angew. Chem. Int. Ed. Engl., 58, 2331-2335. doi: 10.1002/anie.201812628.
- Abstract
- Recently, we observed the first example of a left-handed G-quadruplex structure formed by natural DNA, named Z-G4. We analysed the Z-G4 structure and inspected its primary 28-nt sequence in order to identify motifs that convey the unique left-handed twist. Using circular dichroism spectroscopy, NMR spectroscopy, and X-ray crystallography, we revealed a minimal sequence motif of 12 nt (GTGGTGGTGGTG) for formation of the left-handed DNA G-quadruplex, which is found to be highly abundant in the human genome. A systematic analysis of thymine loop mutations revealed a moderate sequence tolerance, which would further broaden the space of sequences prone to left-handed G-quadruplex formation.
- G4 notes
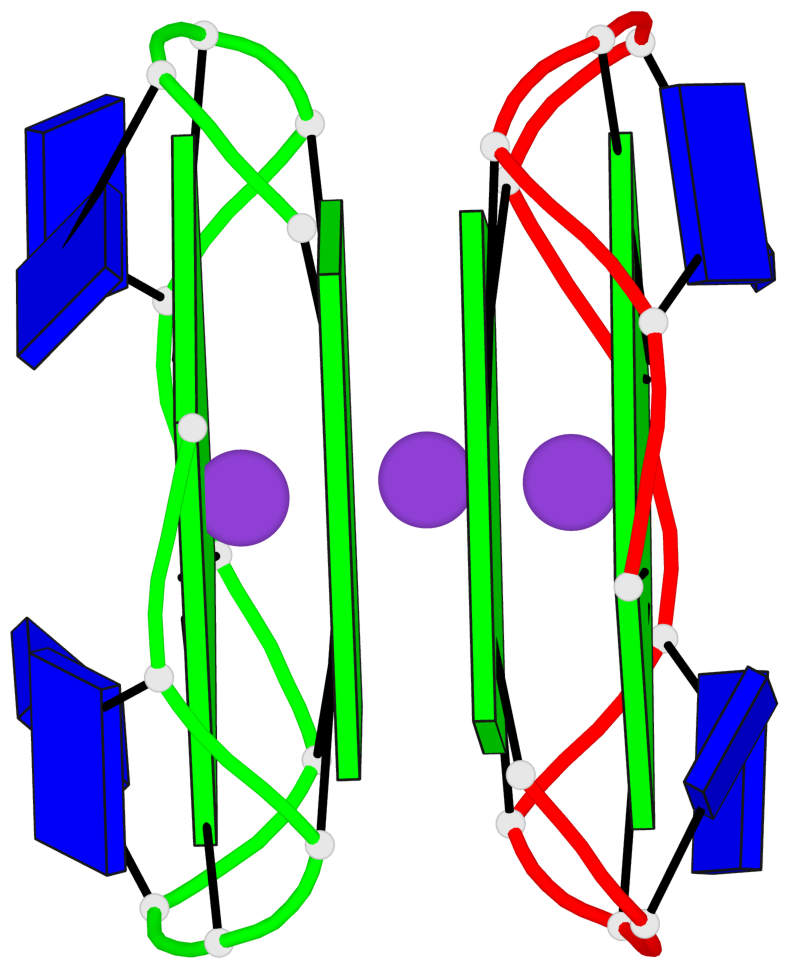
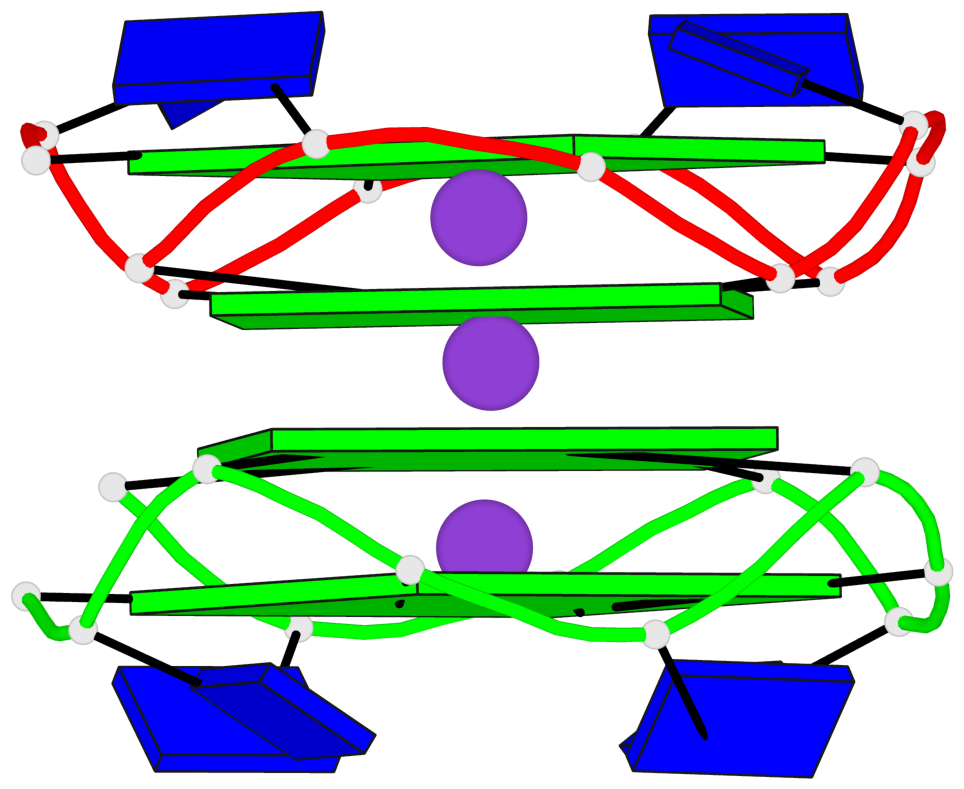
- 4 G-tetrads, 1 G4 helix
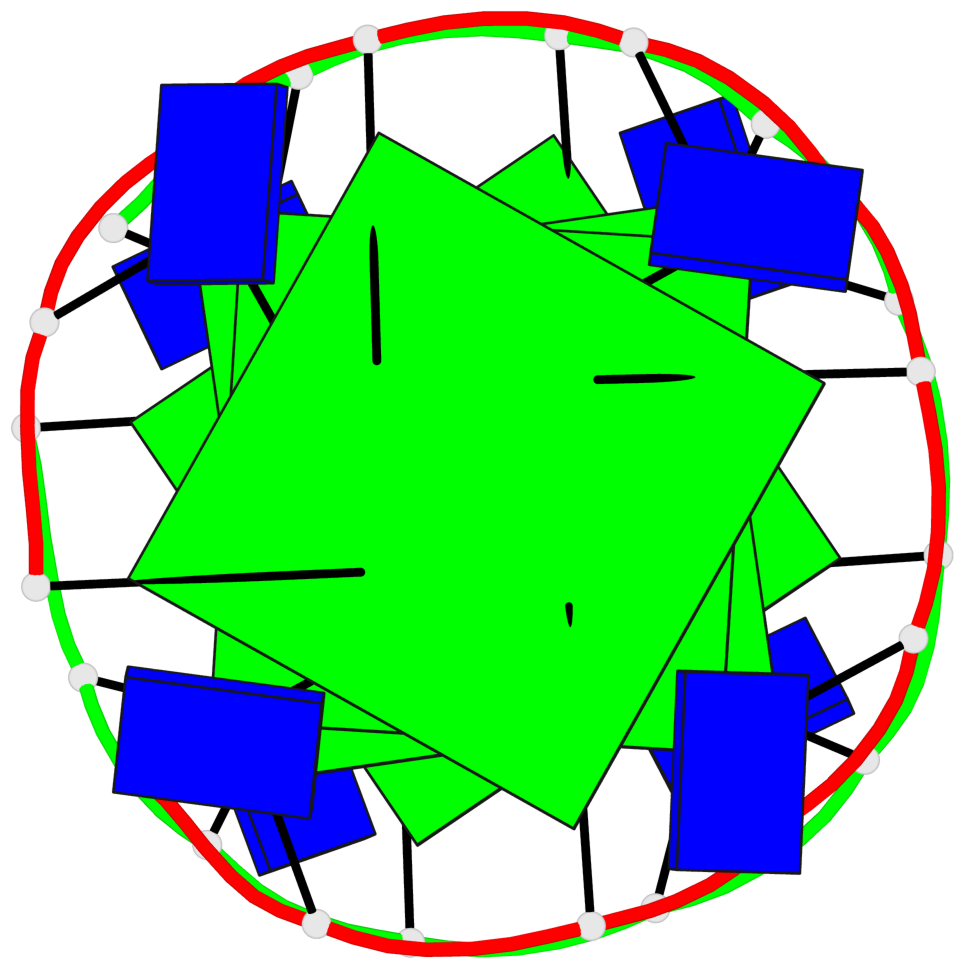
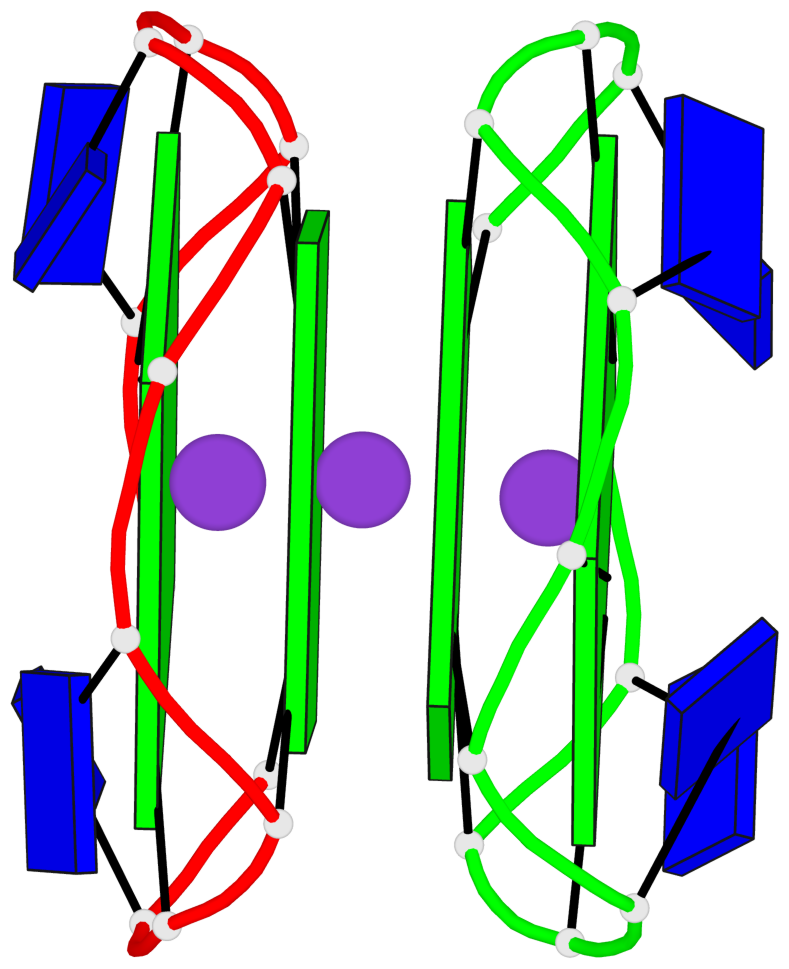
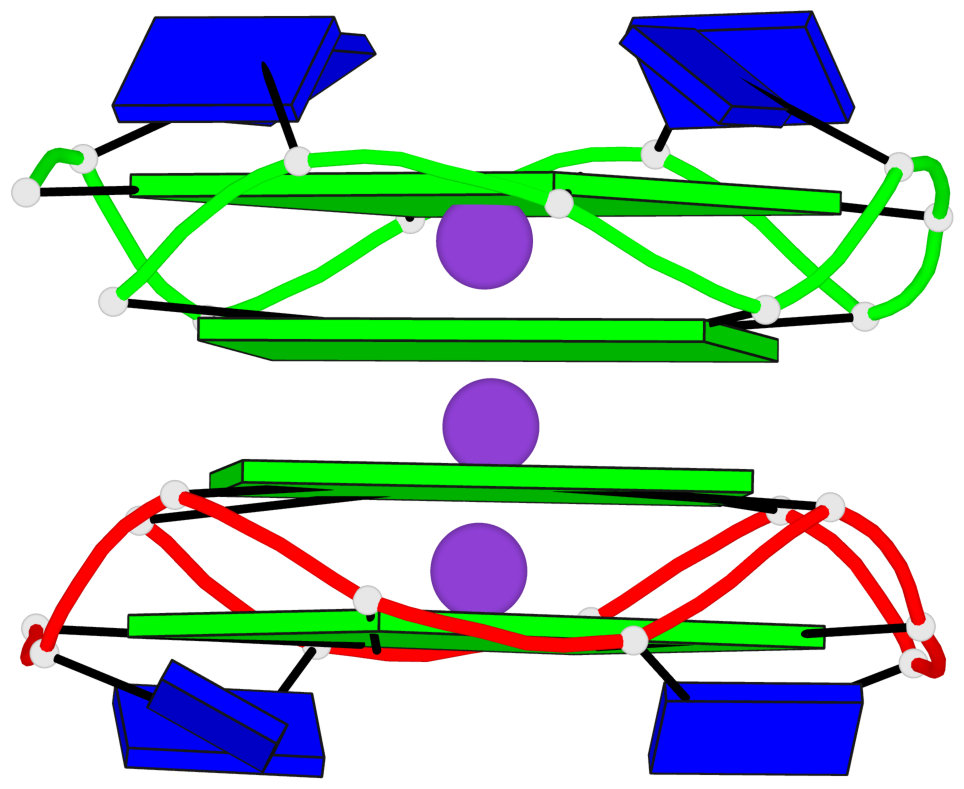
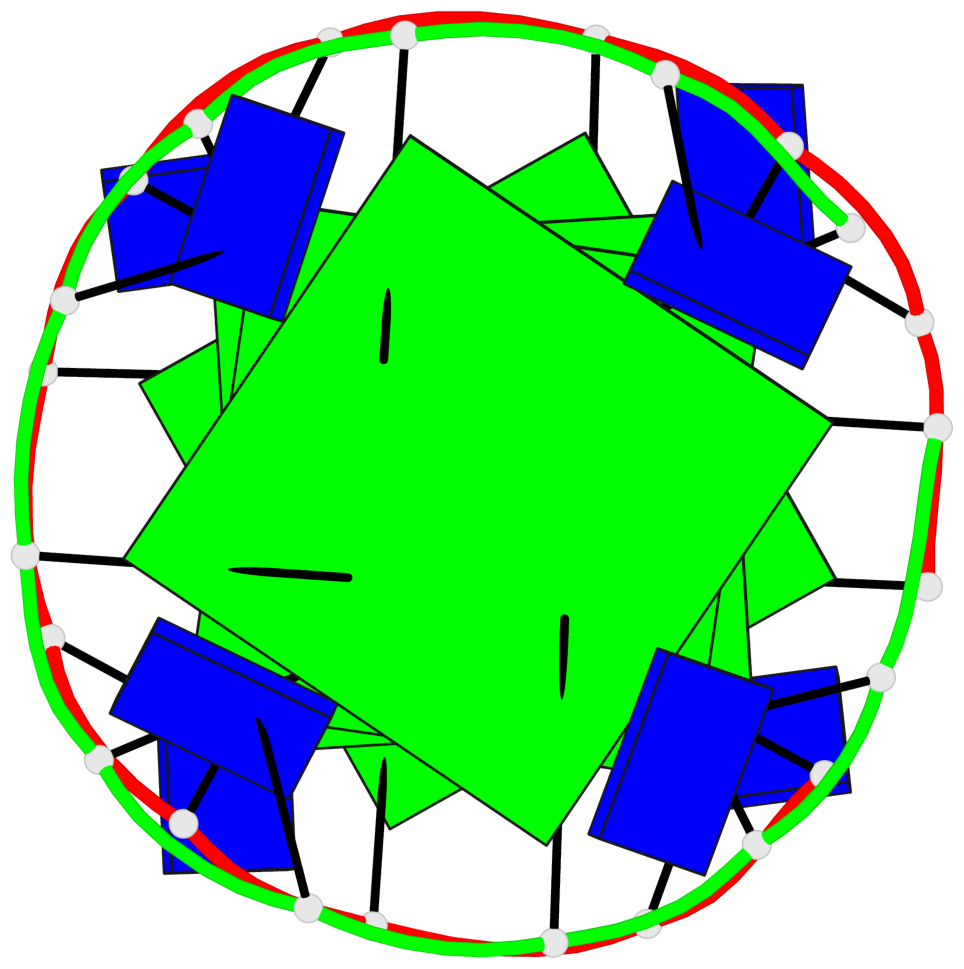
Base-block schematics in six views
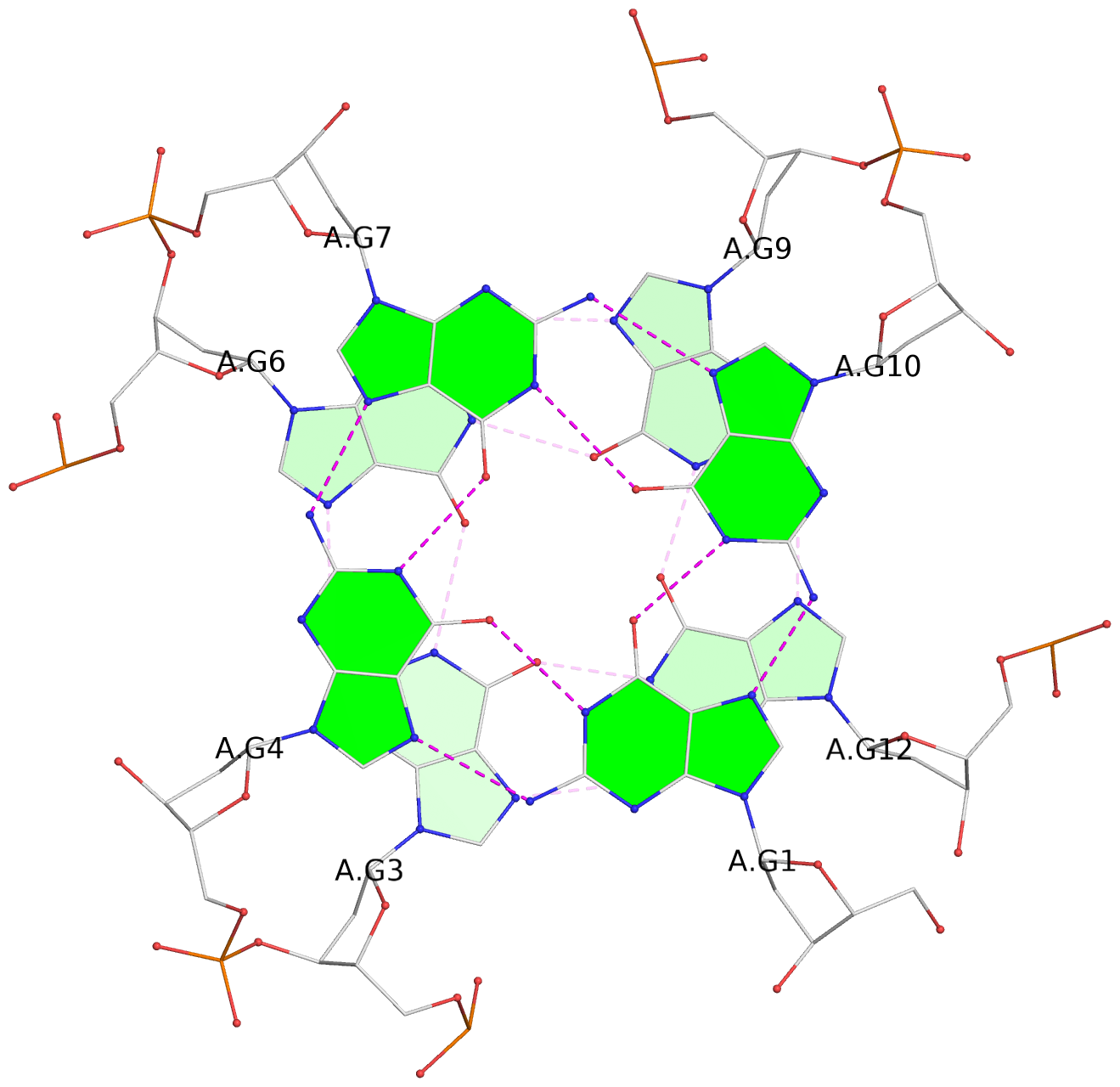
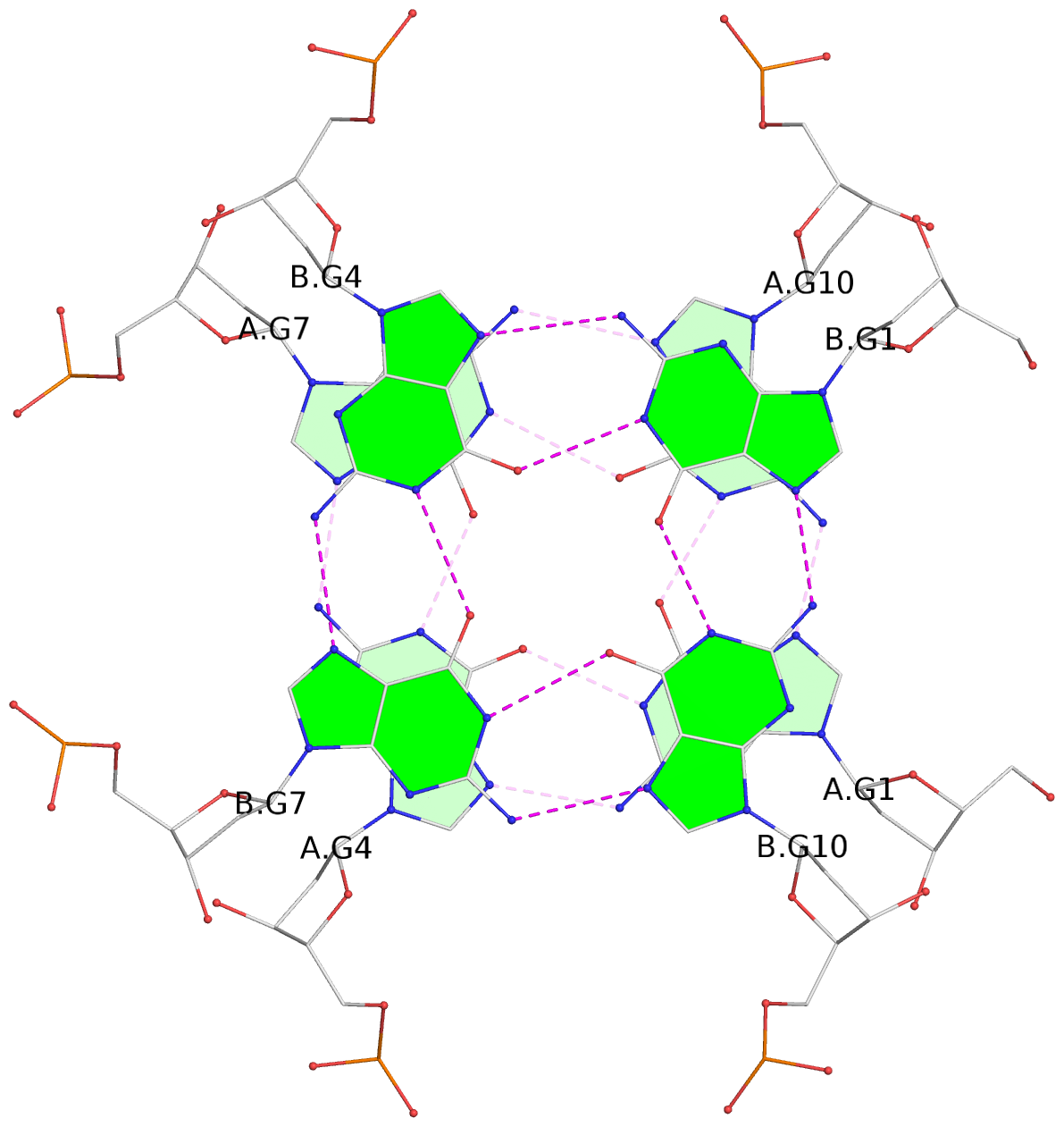
List of 4 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.119 type=planar nts=4 GGGG A.DG1,A.DG4,A.DG7,A.DG10 2 glyco-bond=---- sugar=---- groove=---- planarity=0.312 type=other nts=4 GGGG A.DG3,A.DG6,A.DG9,A.DG12 3 glyco-bond=---- sugar=---- groove=---- planarity=0.114 type=planar nts=4 GGGG B.DG1,B.DG4,B.DG7,B.DG10 4 glyco-bond=---- sugar=---- groove=---- planarity=0.327 type=other nts=4 GGGG B.DG3,B.DG6,B.DG9,B.DG12
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular
List of 4 non-stem G4-loops (including the two closing Gs)
1 type=V-shaped helix=#1 nts=4 GTGG A.DG1,A.DT2,A.DG3,A.DG4 2* type=V-shaped helix=#1 nts=4 GGTG A.DG9,A.DG10,A.DT11,A.DG12 3 type=V-shaped helix=#1 nts=4 GTGG B.DG1,B.DT2,B.DG3,B.DG4 4* type=V-shaped helix=#1 nts=4 GGTG B.DG9,B.DG10,B.DT11,B.DG12