Detailed DSSR results for the G-quadruplex: PDB entry 6ge1
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6ge1
- Class
- RNA
- Method
- NMR
- Summary
- Solution structure of the r(ugguggu)4 RNA quadruplex
- Reference
- Andralojc W, Malgowska M, Sarzynska J, Pasternak K, Szpotkowski K, Kierzek R, Gdaniec Z (2019): "Unraveling the structural basis for the exceptional stability of RNA G-quadruplexes capped by a uridine tetrad at the 3' terminus." RNA, 25, 121-134. doi: 10.1261/rna.068163.118.
- Abstract
- Uridine tetrads (U-tetrads) are a structural element encountered in RNA G-quadruplexes, for example, in the structures formed by the biologically relevant human telomeric repeat RNA. For these molecules, an unexpectedly strong stabilizing influence of a U-tetrad forming at the 3' terminus of a quadruplex was reported. Here we present the high-resolution solution NMR structure of the r(UGGUGGU)4 quadruplex which, in our opinion, provides an explanation for this stabilization. Our structure features a distinctive, abrupt chain reversal just prior to the 3' uridine tetrad. Similar "reversed U-tetrads" were already observed in the crystalline phase. However, our NMR structure coupled with extensive explicit solvent molecular dynamics (MD) simulations identifies some key features of this motif that up to now remained overlooked. These include the presence of an exceptionally stable 2'OH to phosphate hydrogen bond, as well as the formation of an additional K+ binding pocket in the quadruplex groove.
- G4 notes
- 4 G-tetrads, 1 G4 helix, 2 G4 stems, 1 G4 coaxial stack, parallel(4+0), UUUU, coaxial interfaces: 3'/5'
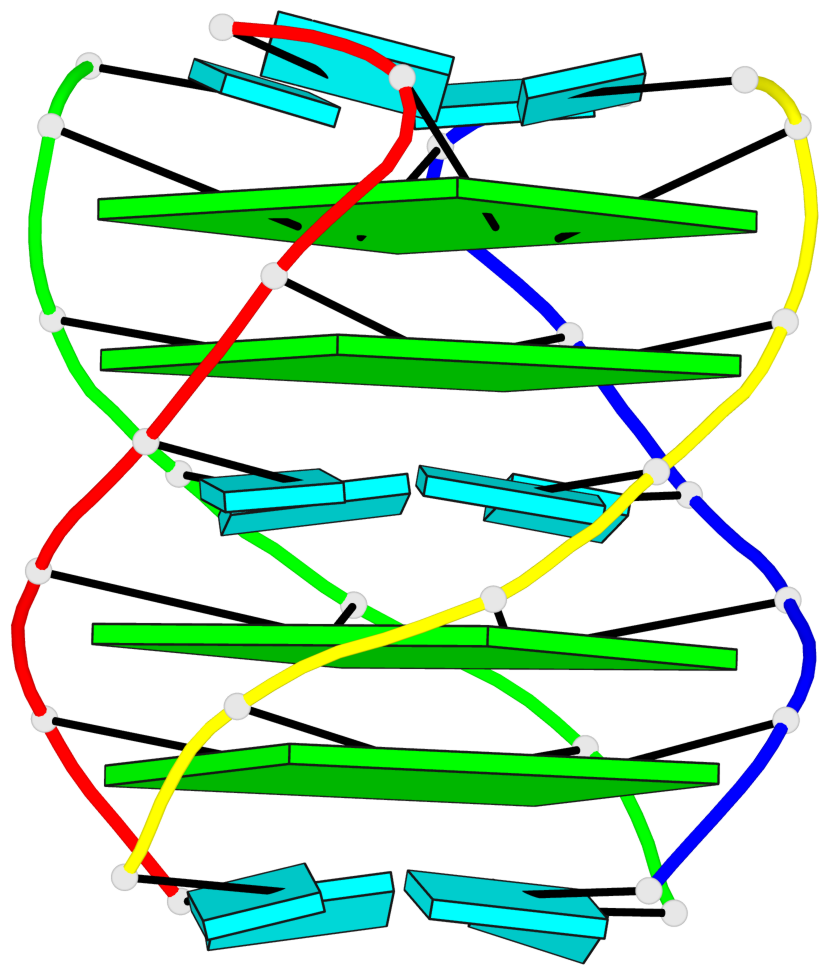
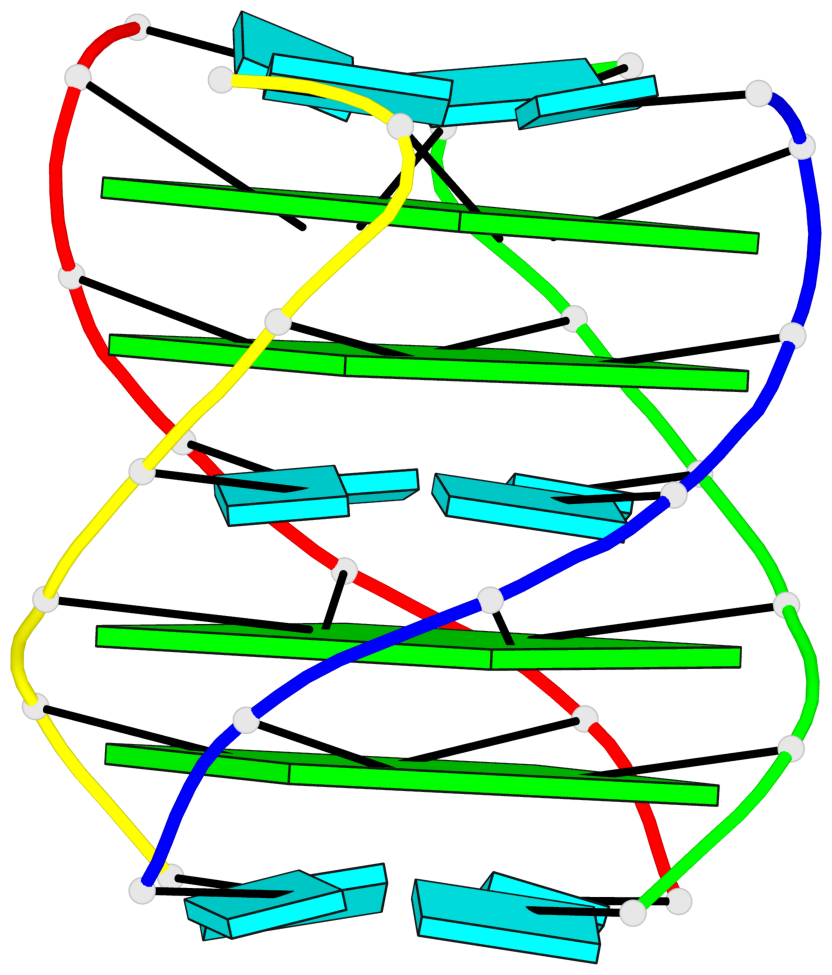
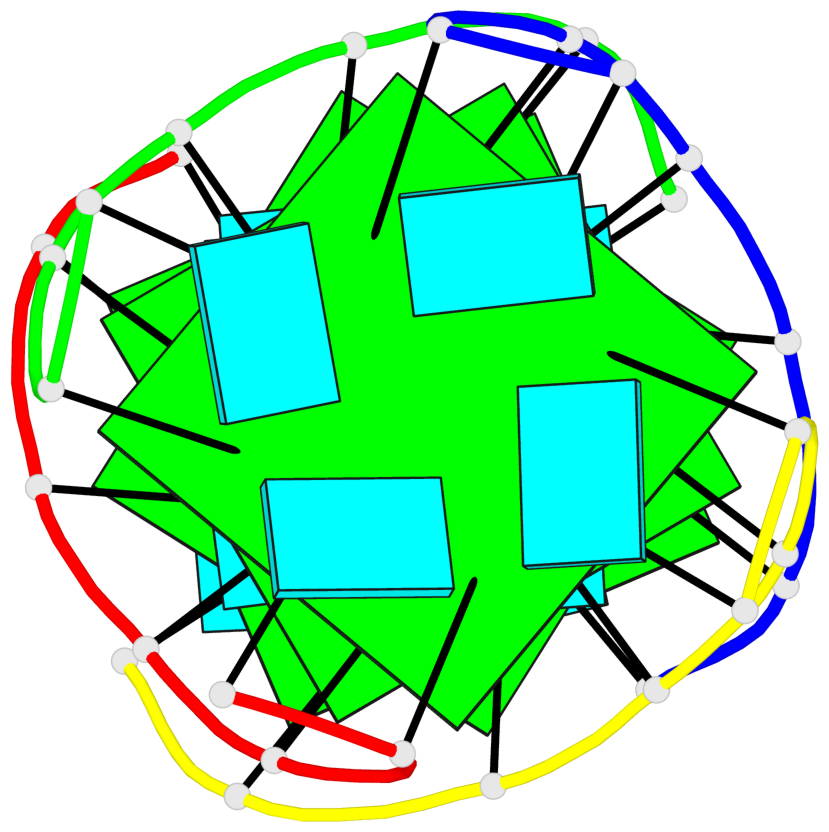
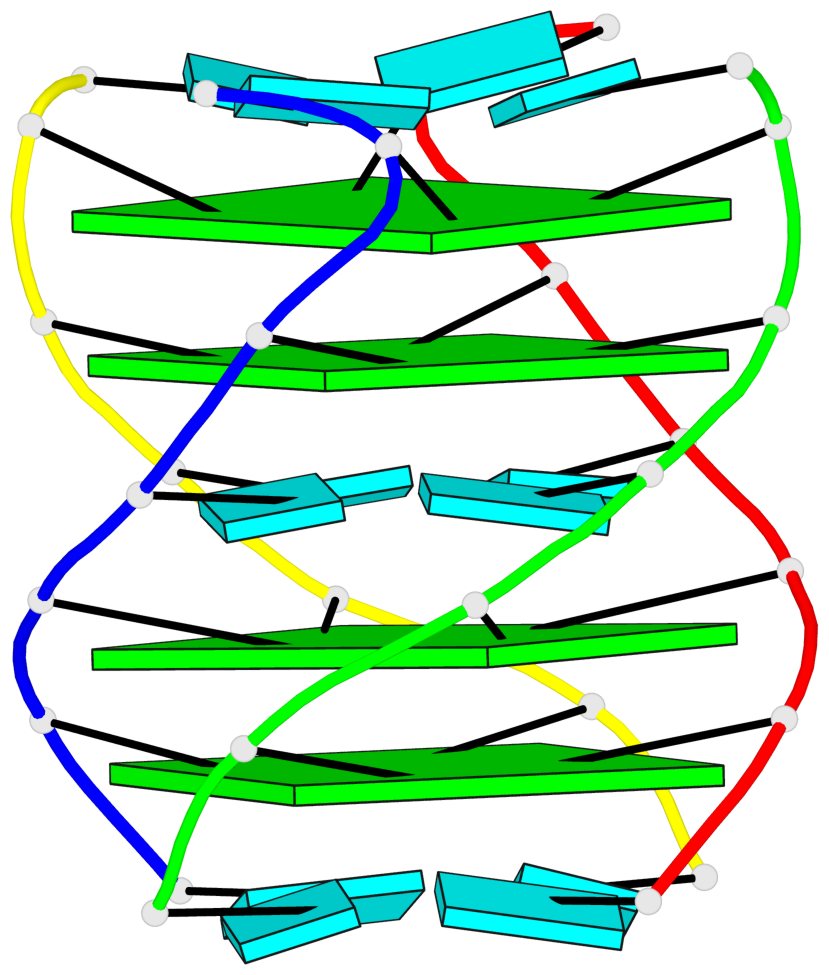
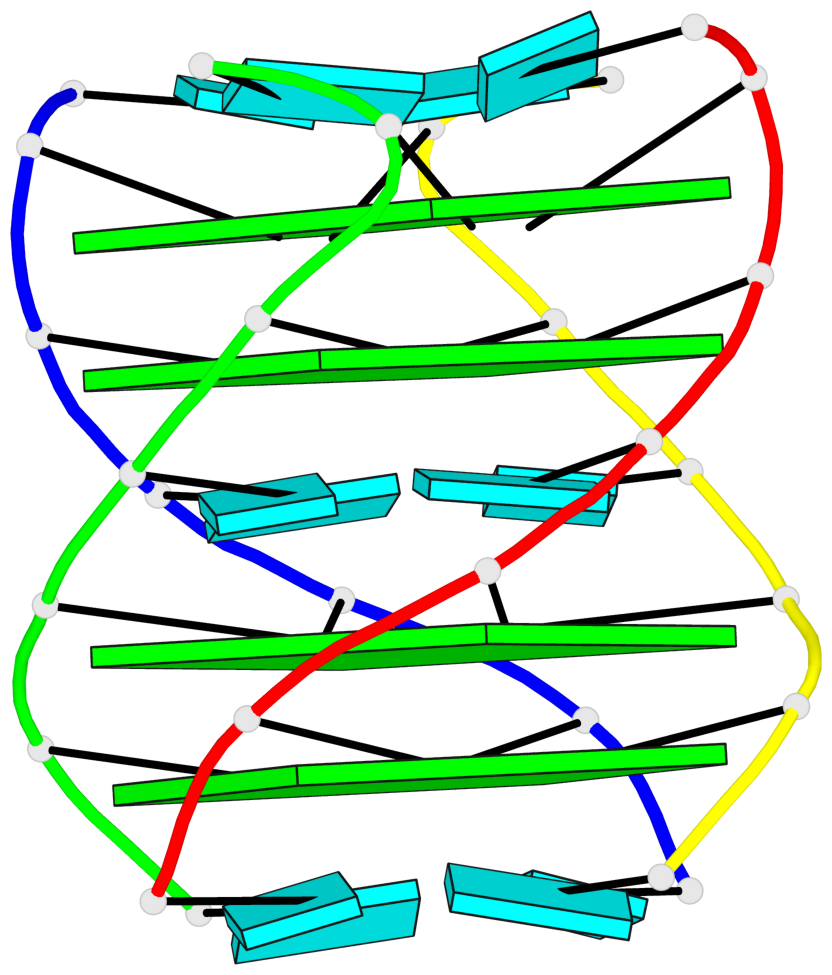
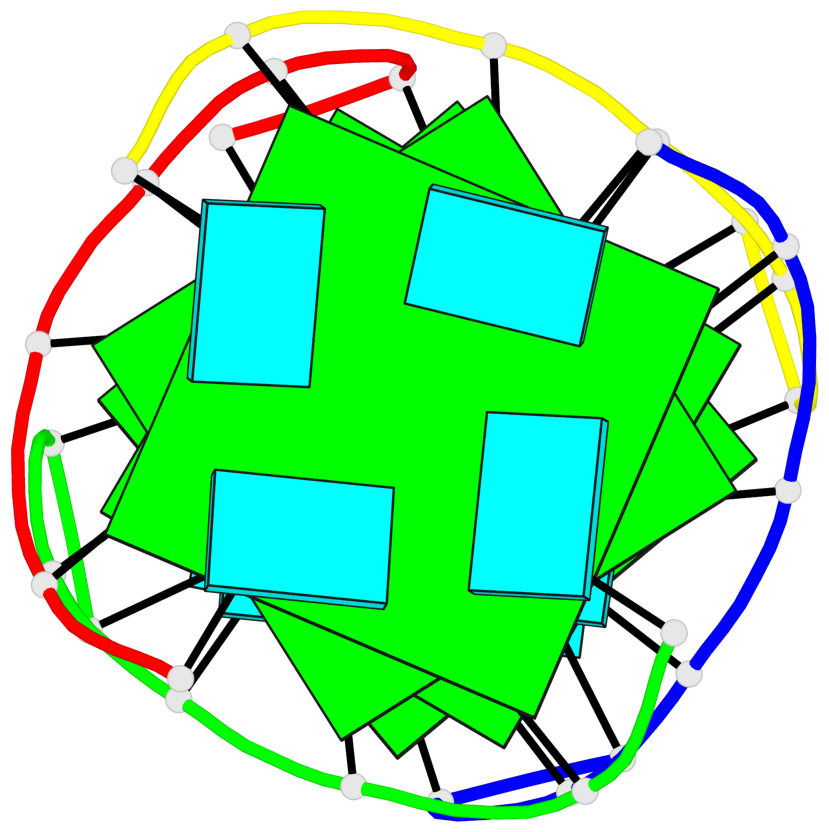
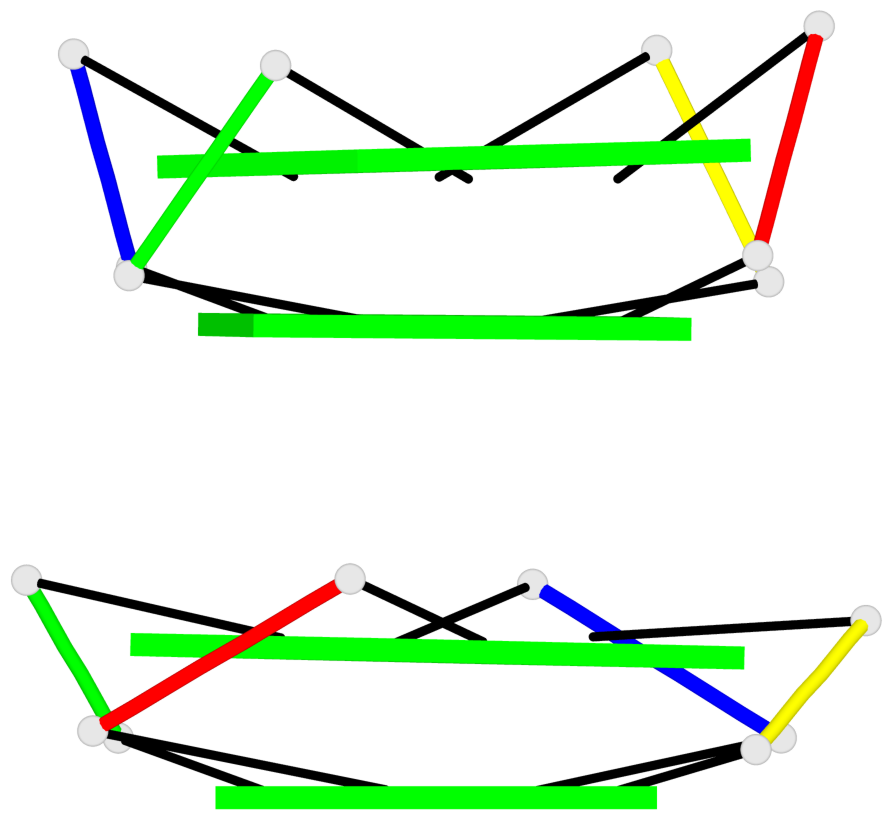
Base-block schematics in six views
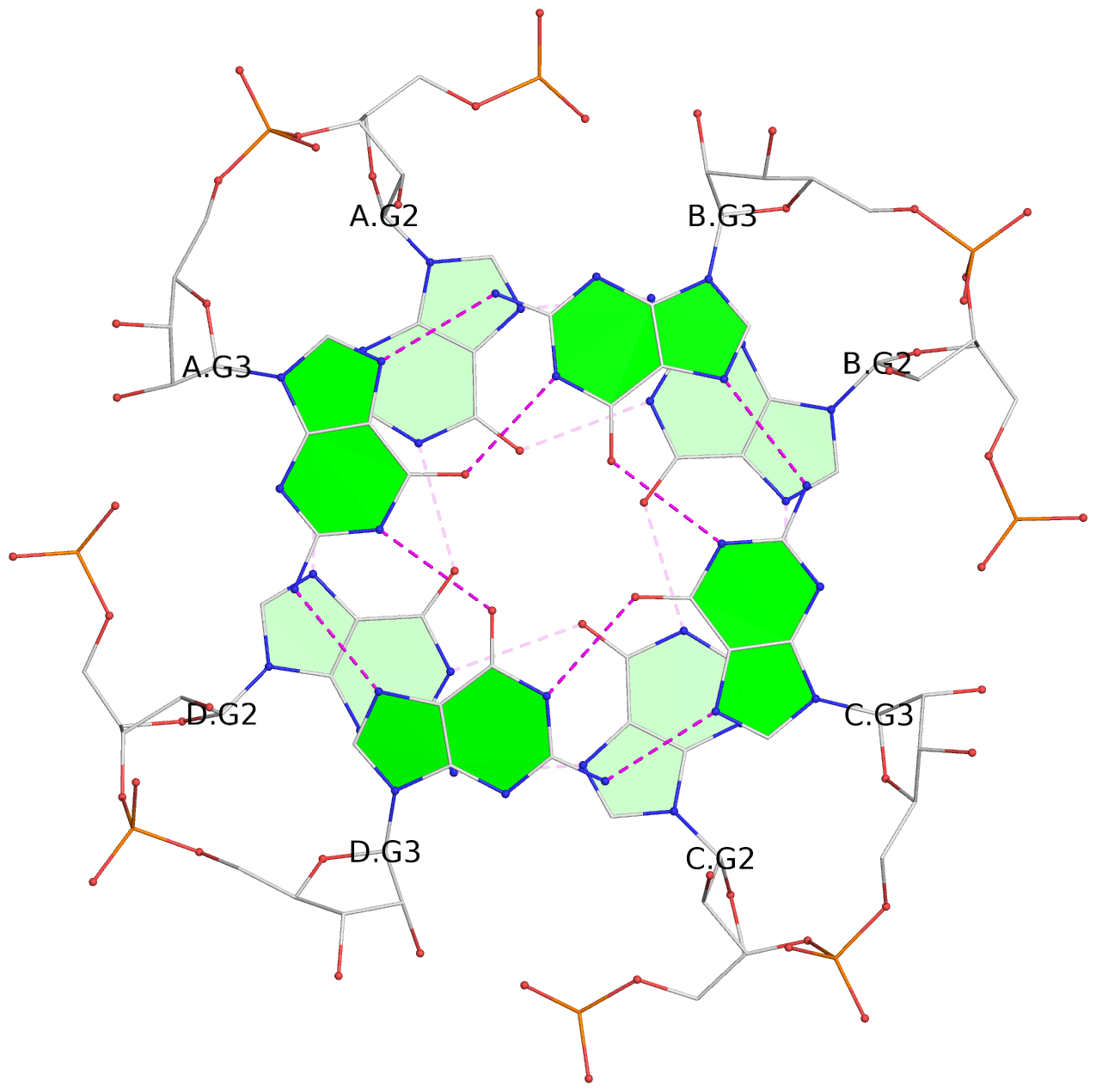
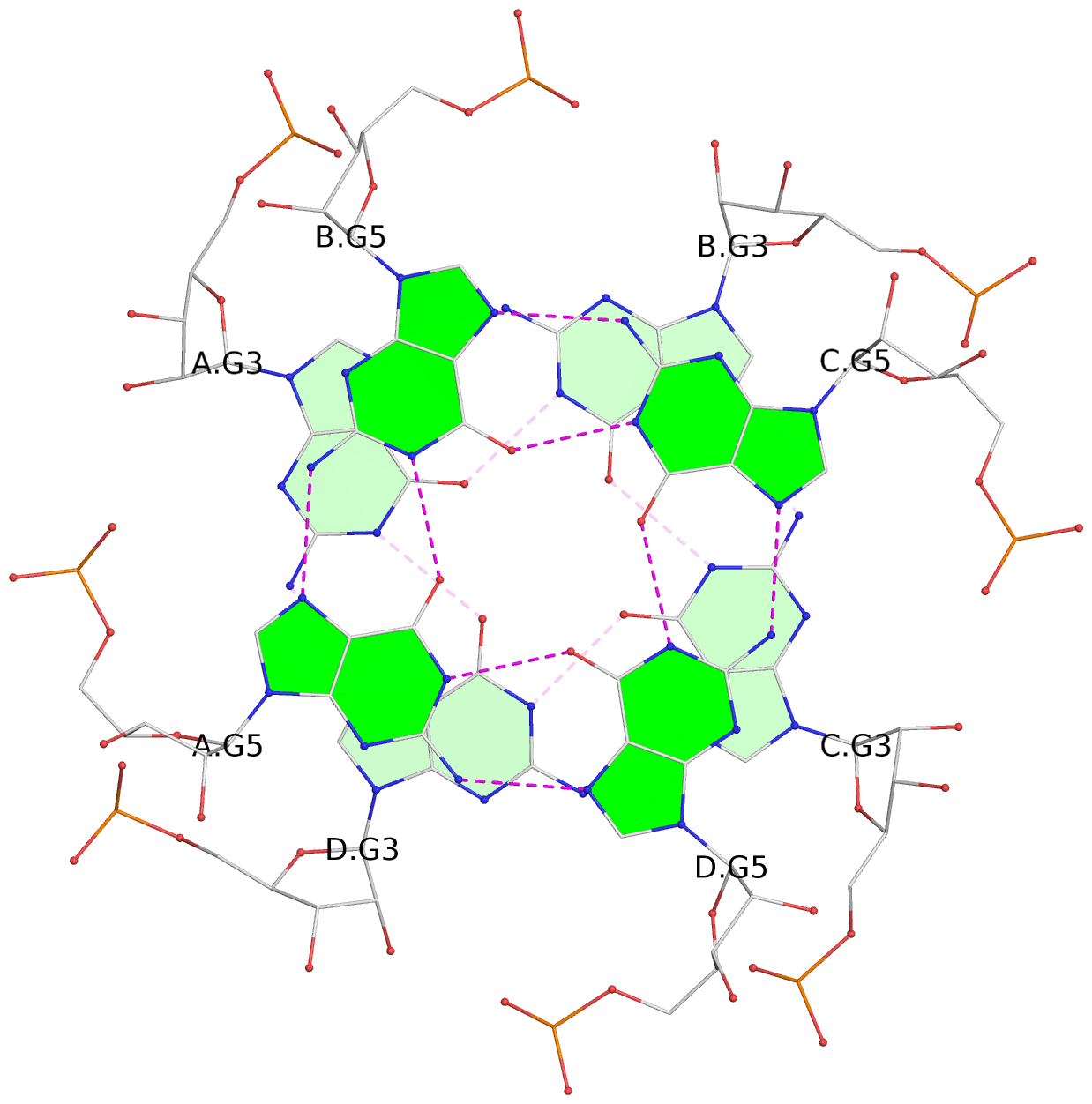
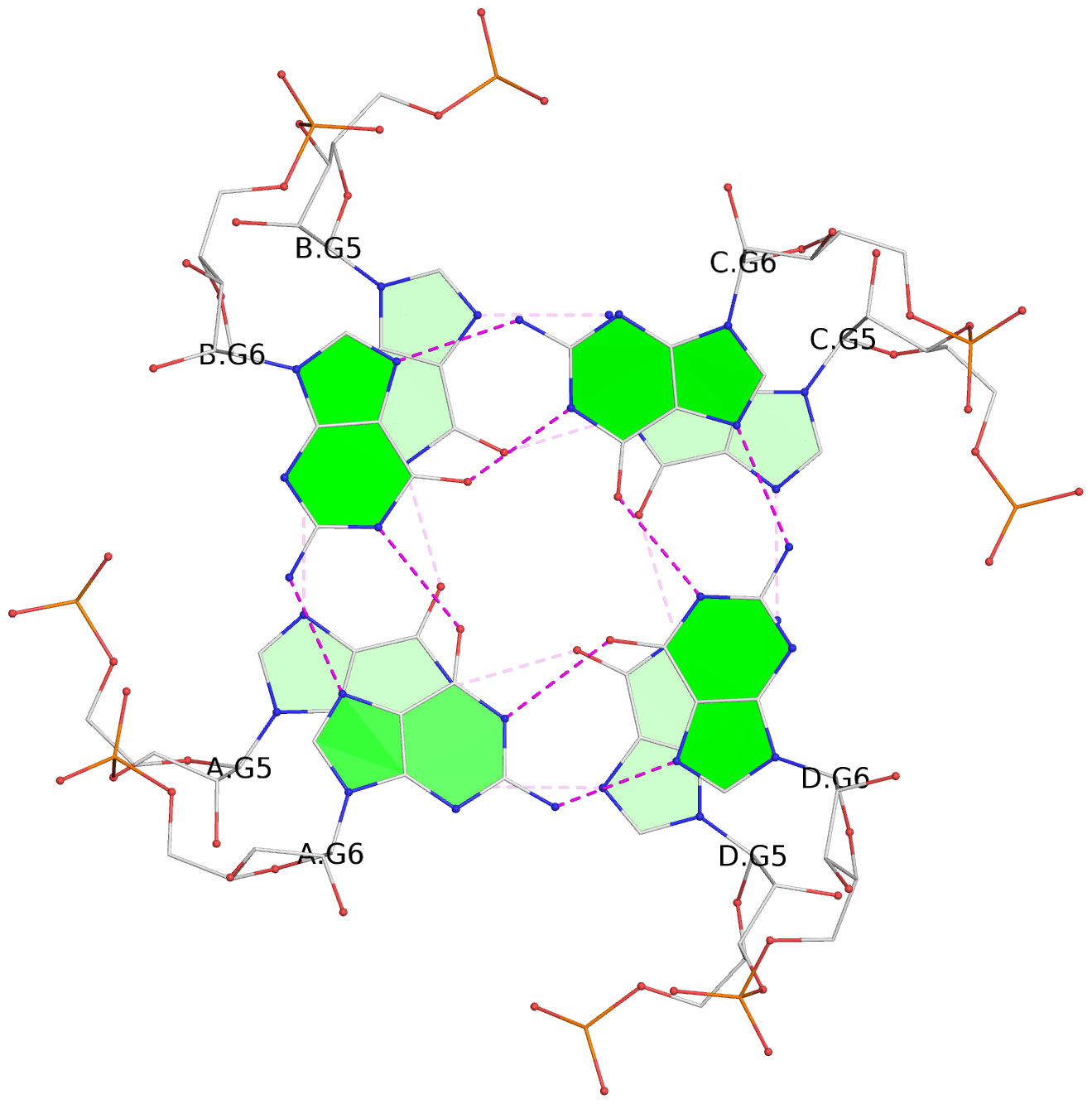
List of 4 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.167 type=other nts=4 GGGG A.G2,D.G2,C.G2,B.G2 2 glyco-bond=---- sugar=3333 groove=---- planarity=0.262 type=other nts=4 GGGG A.G3,D.G3,C.G3,B.G3 3 glyco-bond=---- sugar=3333 groove=---- planarity=0.111 type=planar nts=4 GGGG A.G5,D.G5,C.G5,B.G5 4 glyco-bond=---- sugar=3333 groove=---- planarity=0.368 type=bowl nts=4 GGGG A.G6,D.G6,C.G6,B.G6
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 2 stems
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 0 loops, inter-molecular, UUUU, parallel, parallel(4+0)
Stem#2, 2 G-tetrad layers, 0 loops, inter-molecular, UUUU, parallel, parallel(4+0)
List of 1 G4 coaxial stack
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [3'/5']