Detailed DSSR results for the G-quadruplex: PDB entry 6gh0
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6gh0
- Class
- DNA
- Method
- NMR
- Summary
- Two-quartet kit* g-quadruplex is formed via double-stranded pre-folded structure
- Reference
- Kotar A, Rigo R, Sissi C, Plavec J (2019): "Two-quartet kit* G-quadruplex is formed via double-stranded pre-folded structure." Nucleic Acids Res., 47, 2641-2653. doi: 10.1093/nar/gky1269.
- Abstract
- In the promoter of c-KIT proto-oncogene, whose deregulation has been implicated in many cancers, three G-rich regions (kit1, kit* and kit2) are able to fold into G-quadruplexes. While kit1 and kit2 have been studied in depth, little information is available on kit* folding behavior despite its key role in regulation of c-KIT transcription. Notably, kit* contains consensus sites for SP1 and AP2 transcription factors. Herein, a set of complementary spectroscopic and biophysical methods reveals that kit*, d[GGCGAGGAGGGGCGTGGCCGGC], adopts a chair type antiparallel G-quadruplex with two G-quartets at physiological relevant concentrations of KCl. Heterogeneous ensemble of structures is observed in the presence of Na+ and NH4+ ions, which however stabilize pre-folded structure. In the presence of K+ ions stacking interactions of adenine and thymine residues on the top G-quartet contribute to structural stability together with a G10•C18 base pair and a fold-back motif of the five residues at the 3'-terminal under the bottom G-quartet. The 3'-tail enables formation of a bimolecular pre-folded structure that drives folding of kit* into a single G-quadruplex. Intriguingly, kinetics of kit* G-quadruplex formation matches timescale of transcriptional processes and might demonstrate interplay of kinetic and thermodynamic factors for understanding regulation of c-KIT proto-oncogene expression.
- G4 notes
- 2 G-tetrads, 1 G4 helix, 1 G4 stem, 2(-Lw-Ln-Lw), chair(2+2), UDUD
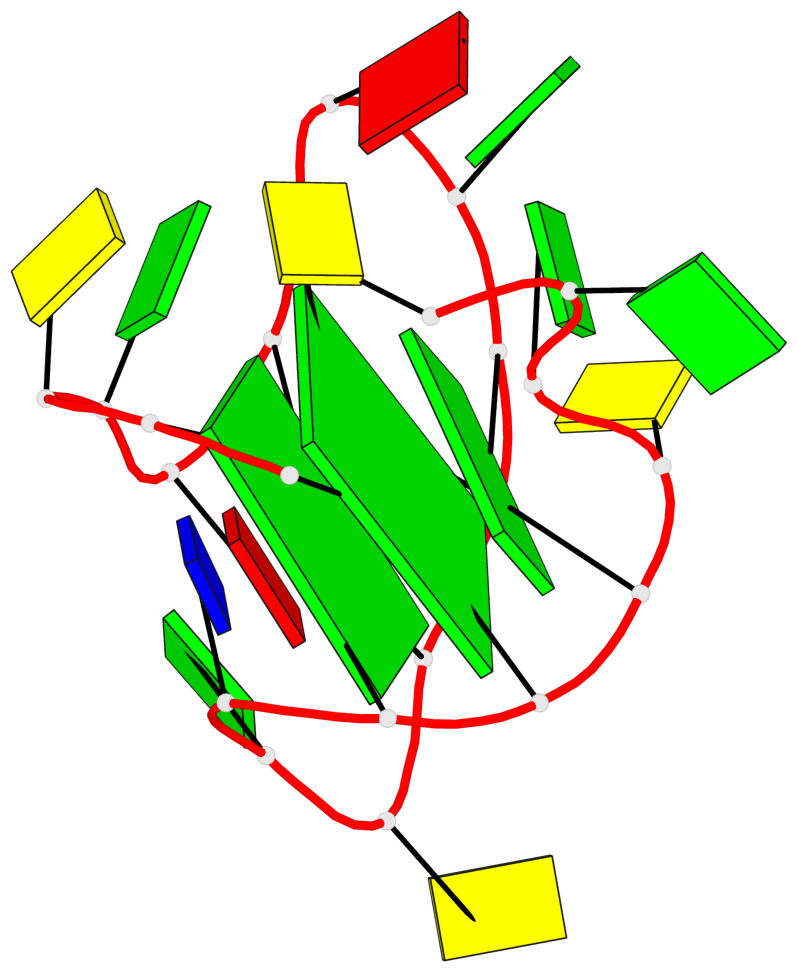
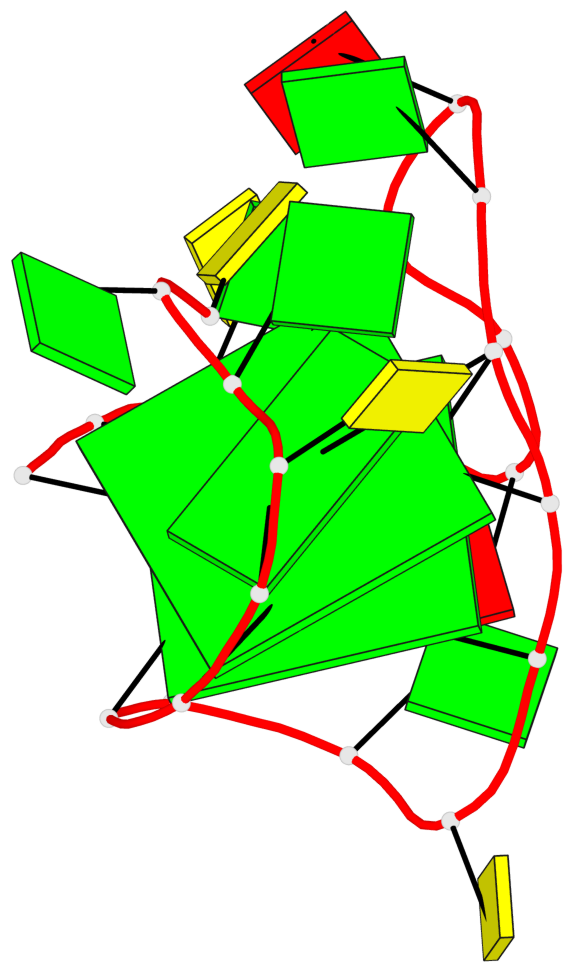
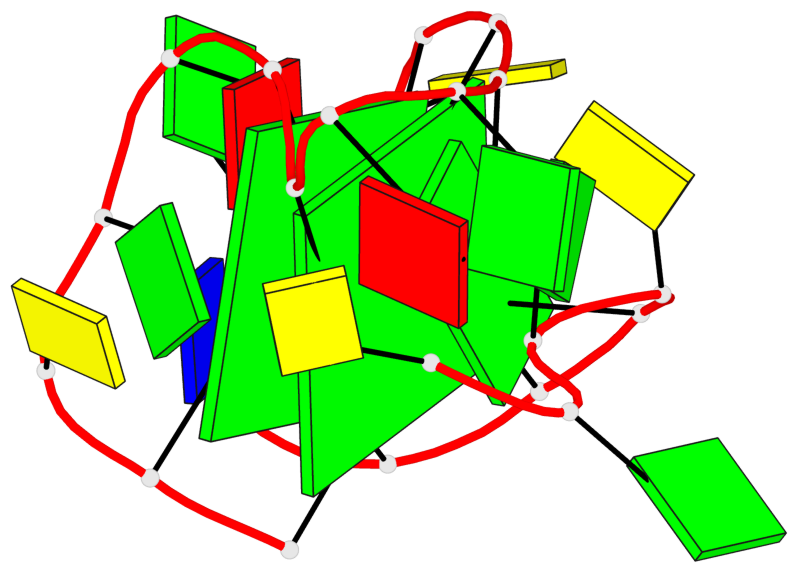
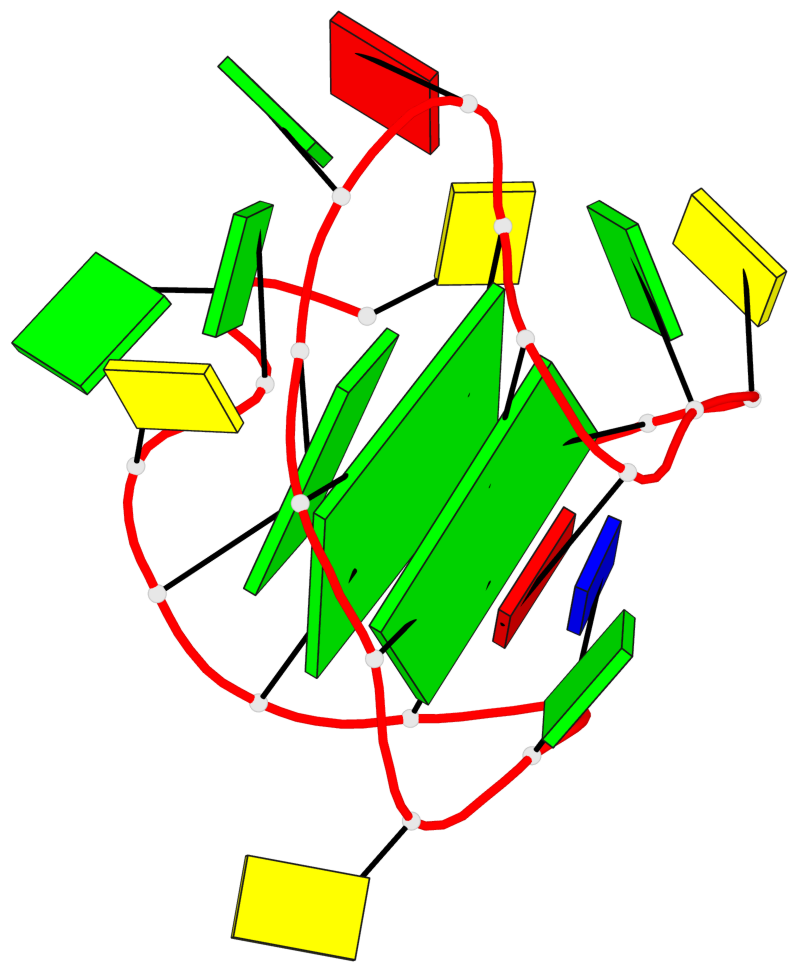
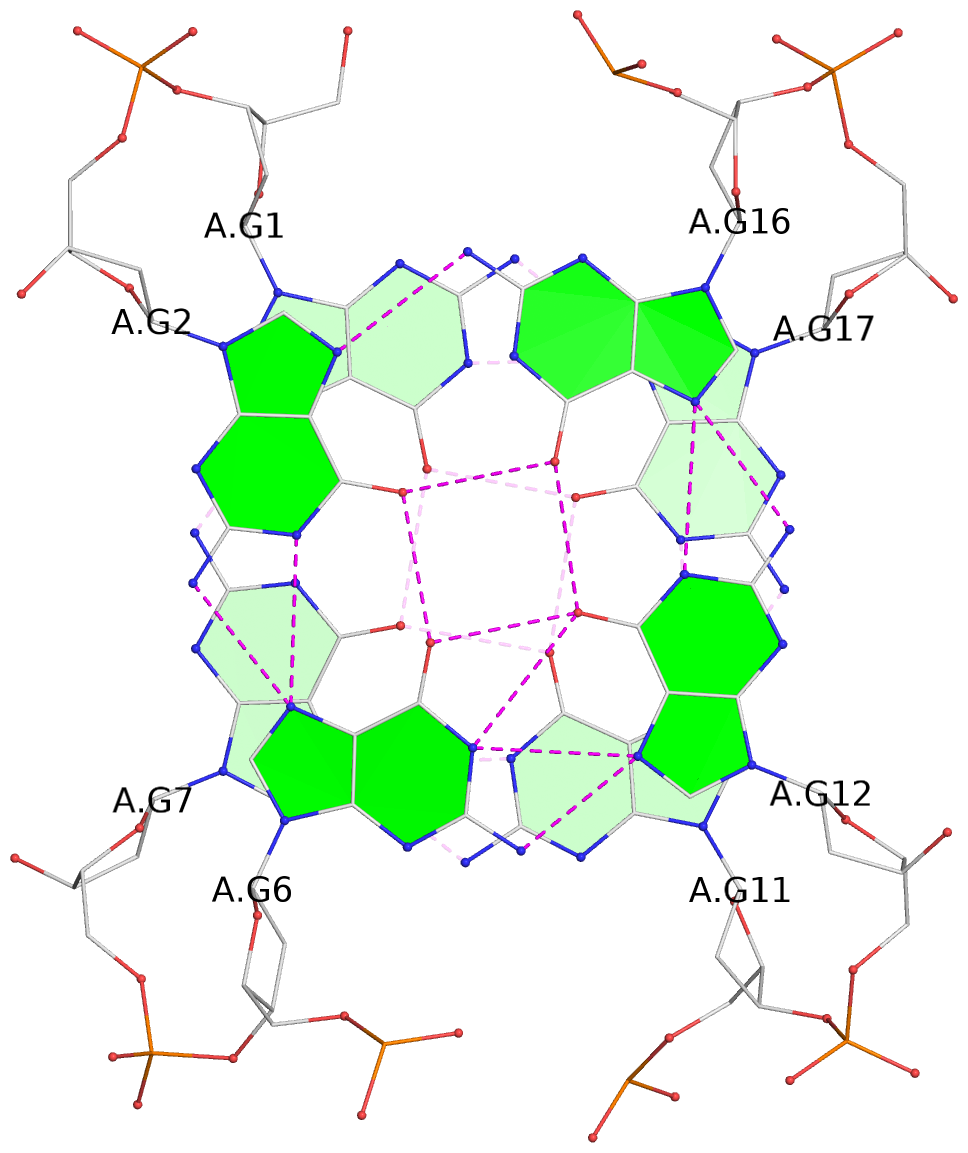
Base-block schematics in six views
List of 2 G-tetrads
1 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.237 type=other nts=4 GGGG A.DG1,A.DG7,A.DG11,A.DG17 2 glyco-bond=-s-s sugar=-.-- groove=wnwn planarity=0.231 type=other nts=4 GGGG A.DG2,A.DG6,A.DG12,A.DG16
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.