Detailed DSSR results for the G-quadruplex: PDB entry 6h1k
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6h1k
- Class
- DNA
- Method
- NMR
- Summary
- The major g-quadruplex form of hiv-1 ltr
- Reference
- Butovskaya E, Heddi B, Bakalar B, Richter SN, Phan AT (2018): "Major G-Quadruplex Form of HIV-1 LTR Reveals a (3 + 1) Folding Topology Containing a Stem-Loop." J. Am. Chem. Soc., 140, 13654-13662. doi: 10.1021/jacs.8b05332.
- Abstract
- Nucleic acids can form noncanonical four-stranded structures called G-quadruplexes. G-quadruplex-forming sequences are found in several genomes including human and viruses. Previous studies showed that the G-rich sequence located in the U3 promoter region of the HIV-1 long terminal repeat (LTR) folds into a set of dynamically interchangeable G-quadruplex structures. G-quadruplexes formed in the LTR could act as silencer elements to regulate viral transcription. Stabilization of LTR G-quadruplexes by G-quadruplex-specific ligands resulted in decreased viral production, suggesting the possibility of targeting viral G-quadruplex structures for antiviral purposes. Among all the G-quadruplexes formed in the LTR sequence, LTR-III was shown to be the major G-quadruplex conformation in vitro. Here we report the NMR structure of LTR-III in K+ solution, revealing the formation of a unique quadruplex-duplex hybrid consisting of a three-layer (3 + 1) G-quadruplex scaffold, a 12-nt diagonal loop containing a conserved duplex-stem, a 3-nt lateral loop, a 1-nt propeller loop, and a V-shaped loop. Our structure showed several distinct features including a quadruplex-duplex junction, representing an attractive motif for drug targeting. The structure solved in this study may be used as a promising target to selectively impair the viral cycle.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 2(D+PX), UD3(1+3), UDDD
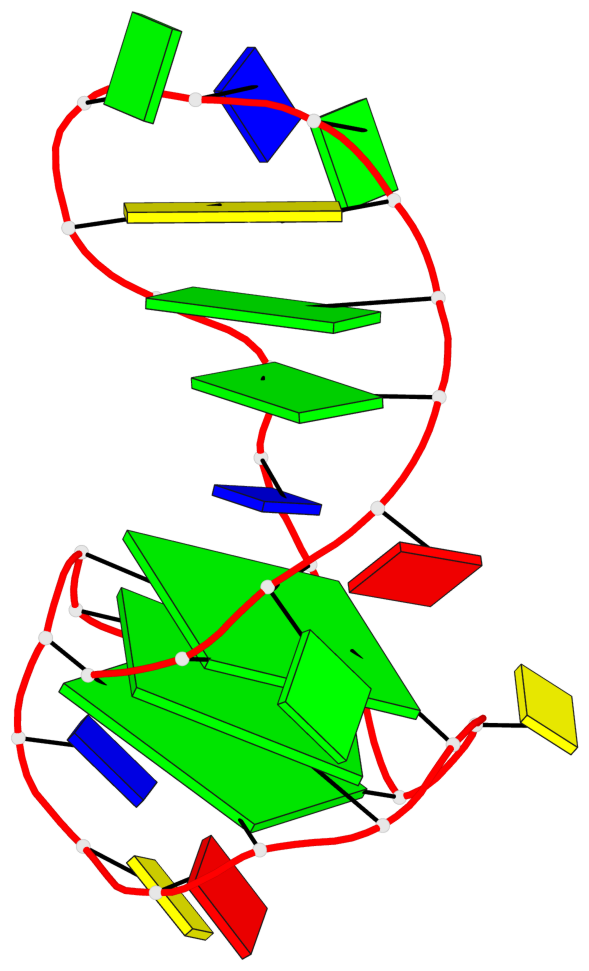
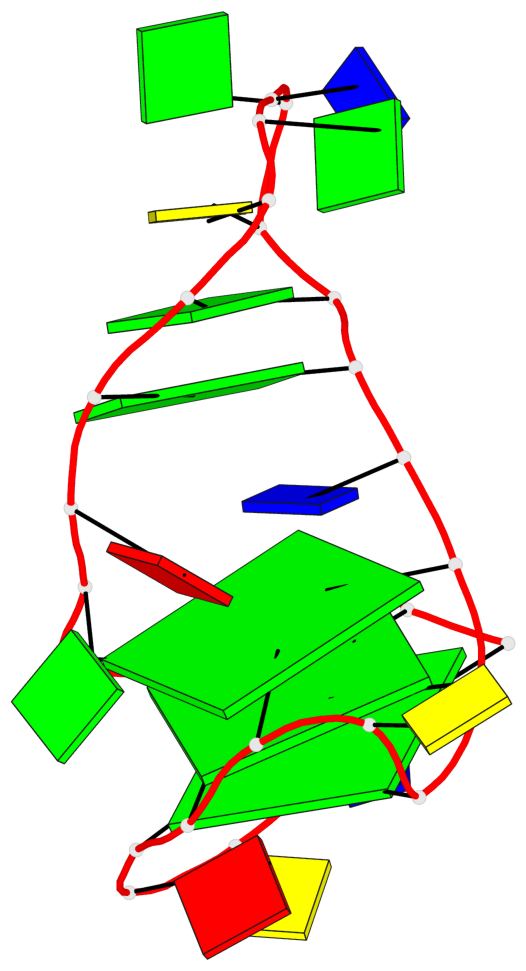
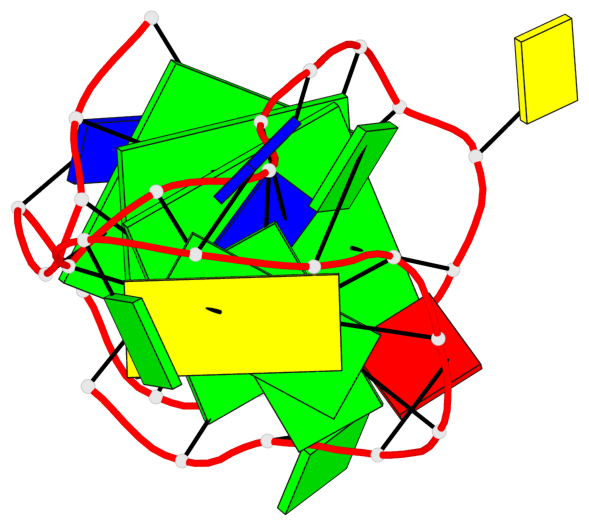
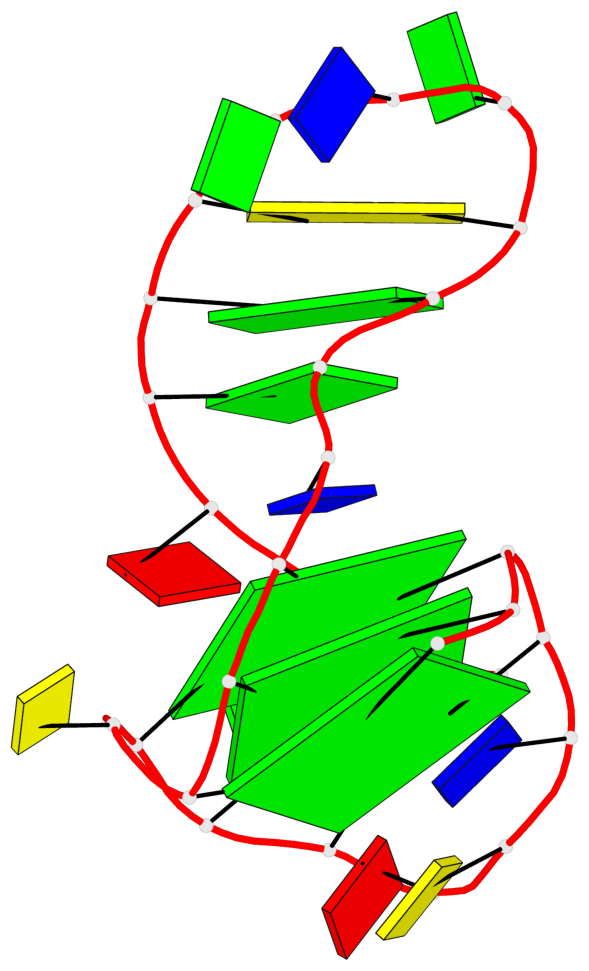
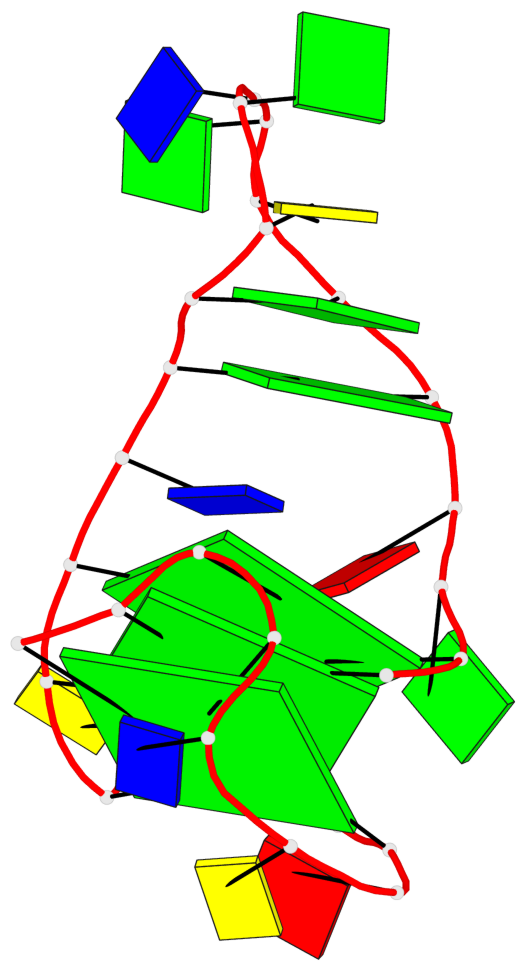
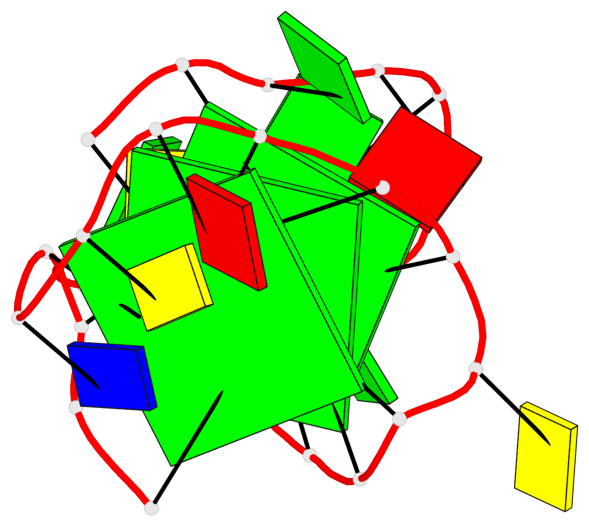
Base-block schematics in six views
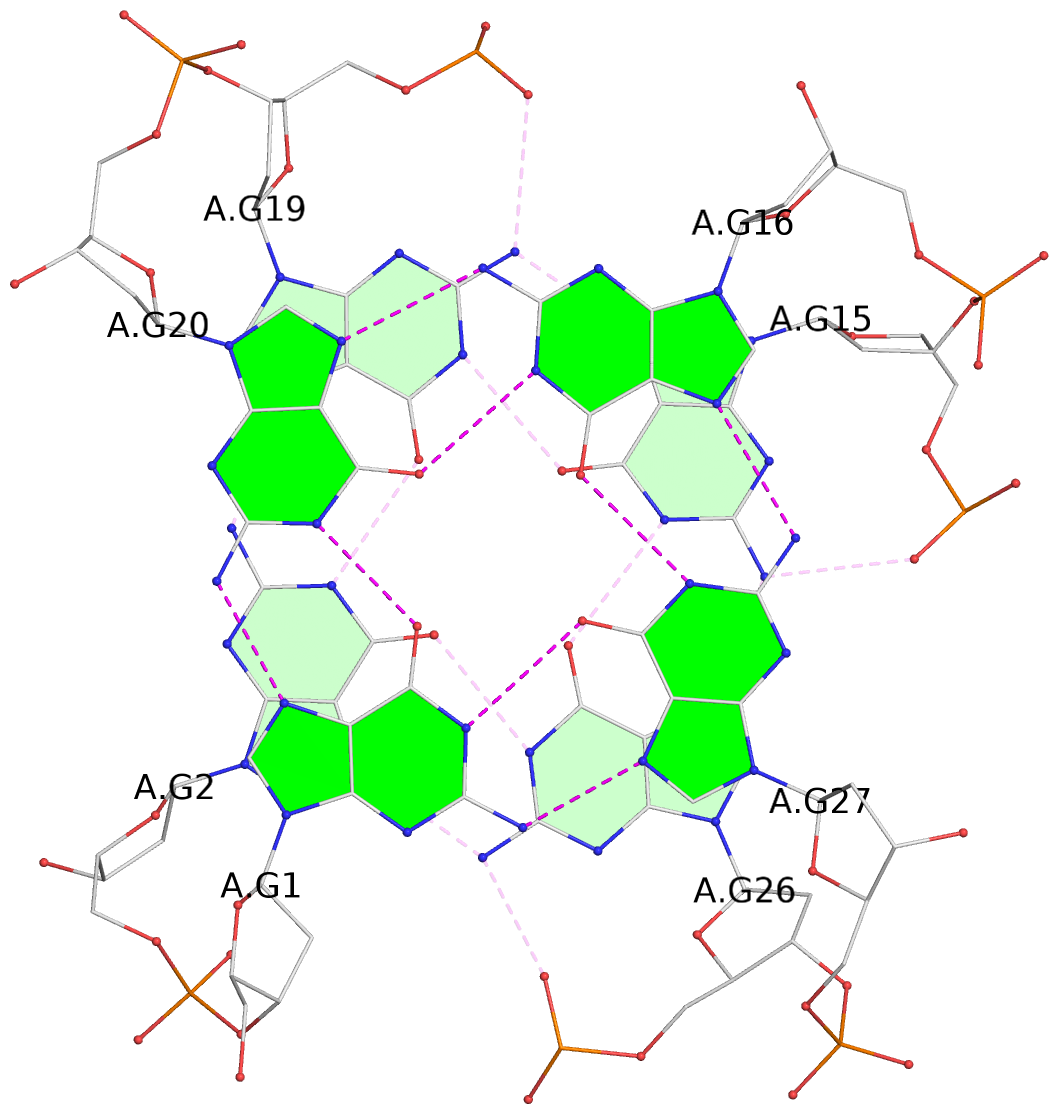
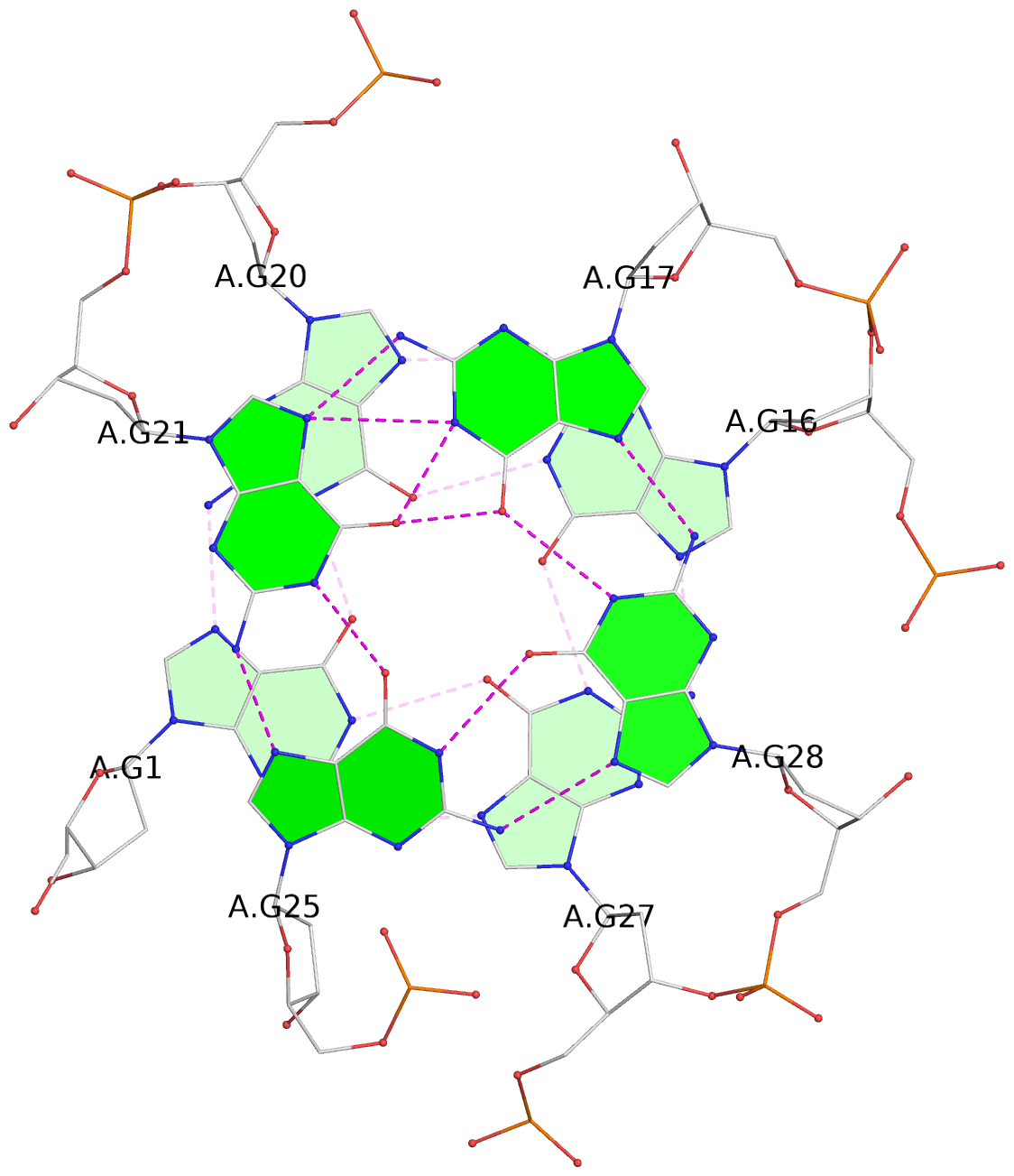
List of 3 G-tetrads
1 glyco-bond=s--- sugar=---- groove=w--n planarity=0.149 type=planar nts=4 GGGG A.DG1,A.DG20,A.DG16,A.DG27 2 glyco-bond=-sss sugar=.-.3 groove=w--n planarity=0.136 type=planar nts=4 GGGG A.DG2,A.DG19,A.DG15,A.DG26 3 glyco-bond=--s- sugar=--.- groove=-wn- planarity=0.307 type=other nts=4 GGGG A.DG17,A.DG21,A.DG25,A.DG28
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UDDD, hybrid-(mixed), 2(D+PX), UD3(1+3)
List of 2 non-stem G4-loops (including the two closing Gs)
1 type=lateral helix=#1 nts=5 GACTG A.DG21,A.DA22,A.DC23,A.DT24,A.DG25 2 type=V-shaped helix=#1 nts=4 GGGG A.DG25,A.DG26,A.DG27,A.DG28