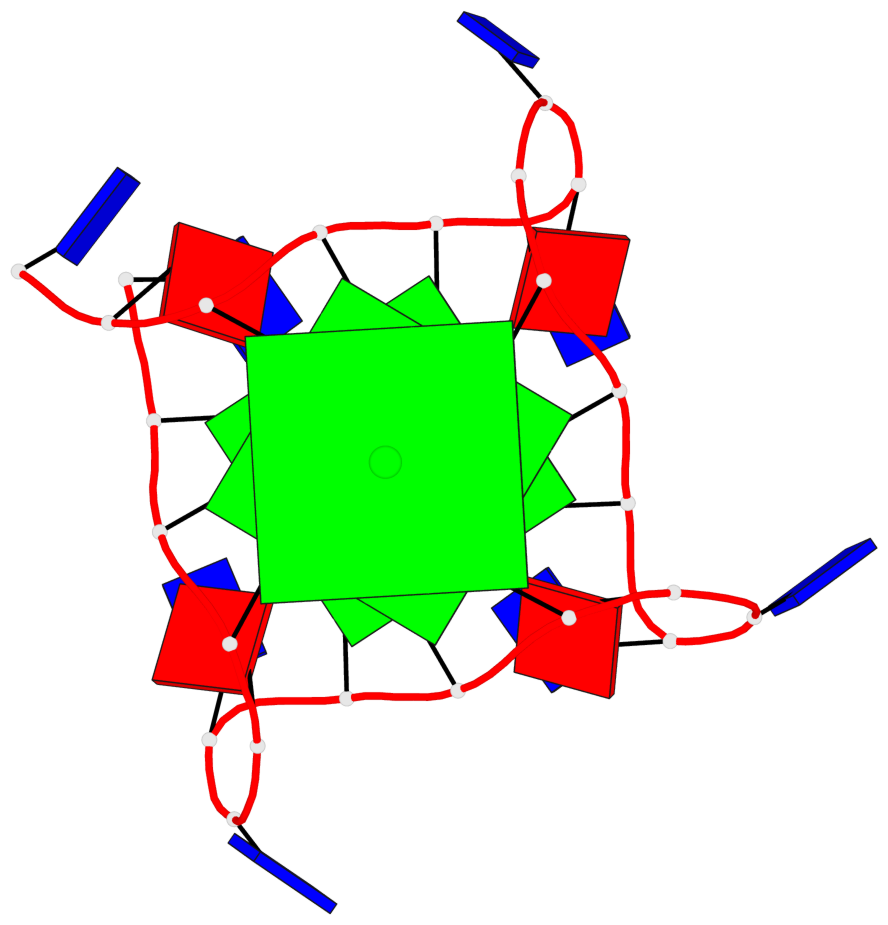
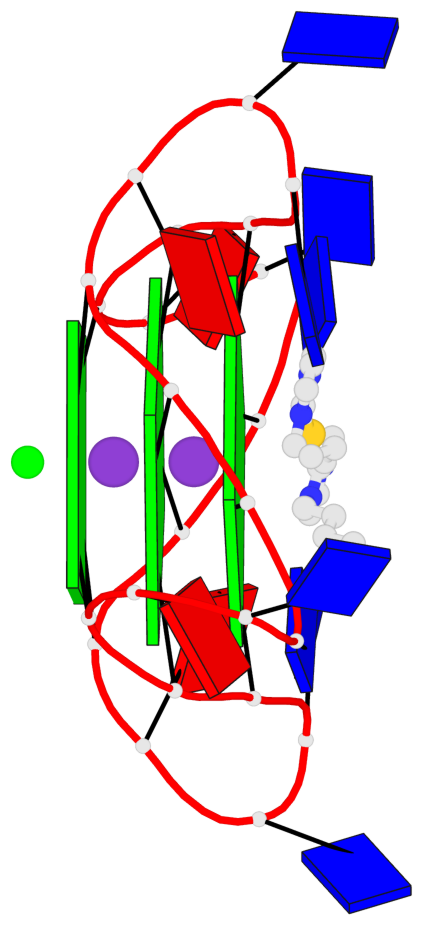
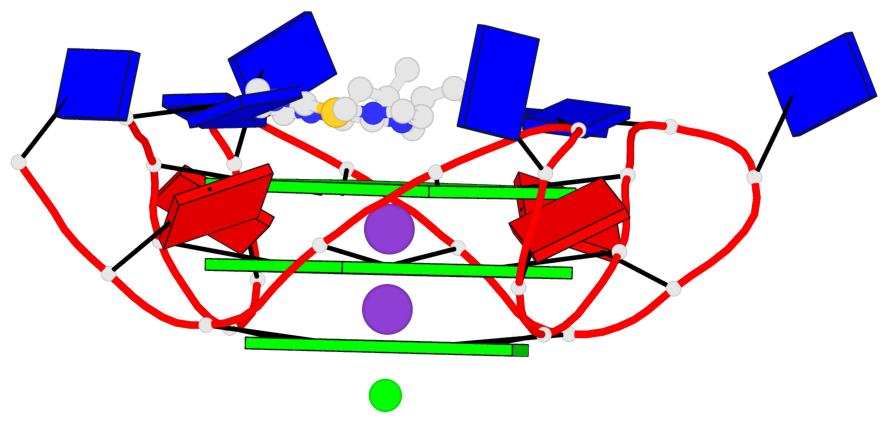
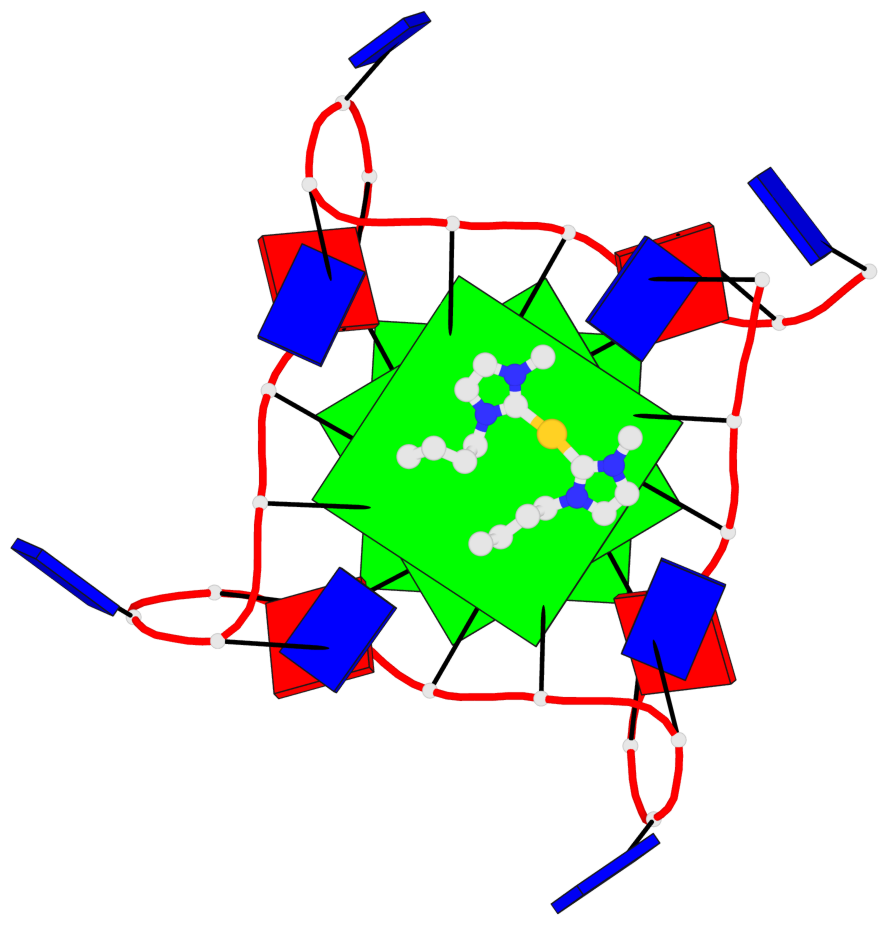
Detailed DSSR results for the G-quadruplex: PDB entry 6h5r
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6h5r
- Class
- DNA
- Method
- X-ray (2.0 Å)
- Summary
- Structure of the complex of a human telomeric DNA with bis(1-butyl-3-methyl-imidazole-2-ylidene) gold(i)
- Reference
- Guarra F, Marzo T, Ferraroni M, Papi F, Bazzicalupi C, Gratteri P, Pescitelli G, Messori L, Biver T, Gabbiani C (2018): "Interaction of a gold(i) dicarbene anticancer drug with human telomeric DNA G-quadruplex: solution and computationally aided X-ray diffraction analysis." Dalton Trans, 47, 16132-16138. doi: 10.1039/c8dt03607a.
- Abstract
- The bis carbene gold(i) complex [Au(1-butyl-3-methyl-2-ylidene)2]PF6, ([Au(NHC)2]PF6 hereafter), holds remarkable interest as a perspective anticancer agent. The compound is stable under physiological like conditions: its original structure is retained even in the presence of excess glutathione (GSH). Previous studies revealed its high cytotoxicity in vitro that correlates with the impairment of crucial metabolic and enzymatic cellular processes (Magherini et al., Oncotarget, 2018, 9, 28042). Here, the interaction of [Au(NHC)2]PF6 with the human telomeric DNA G-quadruplex Tel23 has been investigated in solution by means of high resolution mass spectrometry. ESI MS experiments well document the formation of stable 1 : 1 adducts between the biscarbene gold complex - in its intact form - and the DNA G-quadruplex Tel23. Next, through independent biophysical methods, we show that [Au(NHC)2]PF6 binding does not significantly affect the G quadruplex melting temperature nor its conformation. The crystal structure for the [Au(NHC)2]+/Tel24 adduct was eventually determined by a joint X-ray diffraction and in silico simulation approach. Through the careful integration of solution and solid-state data, a quite clear picture emerges for the interaction of this gold complex with the Tel23 G-quadruplex.
- G4 notes
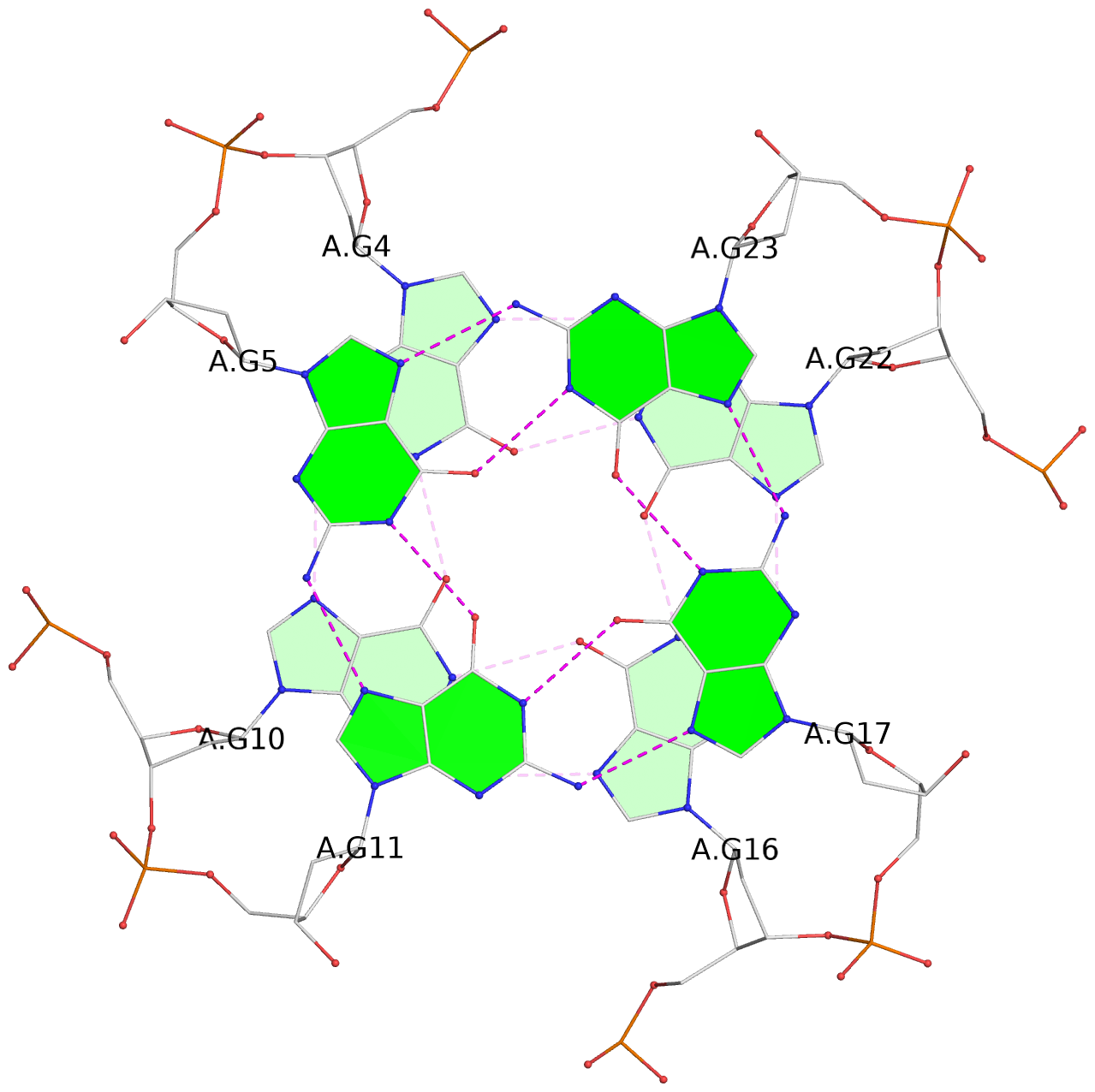
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
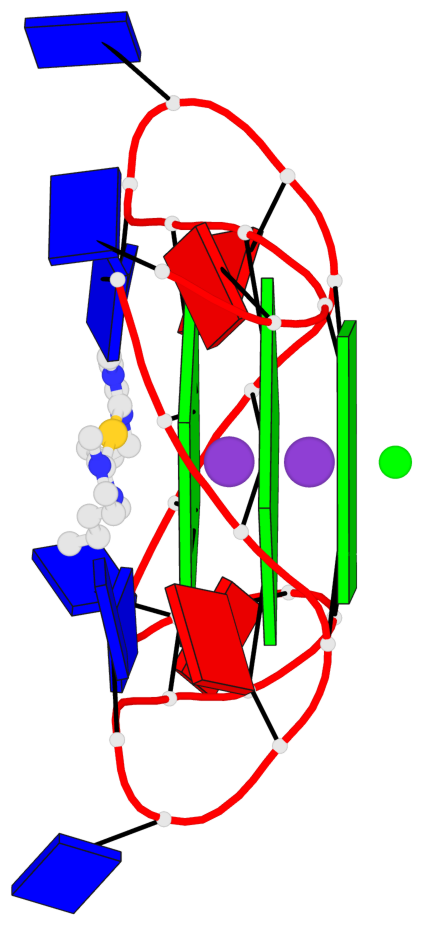
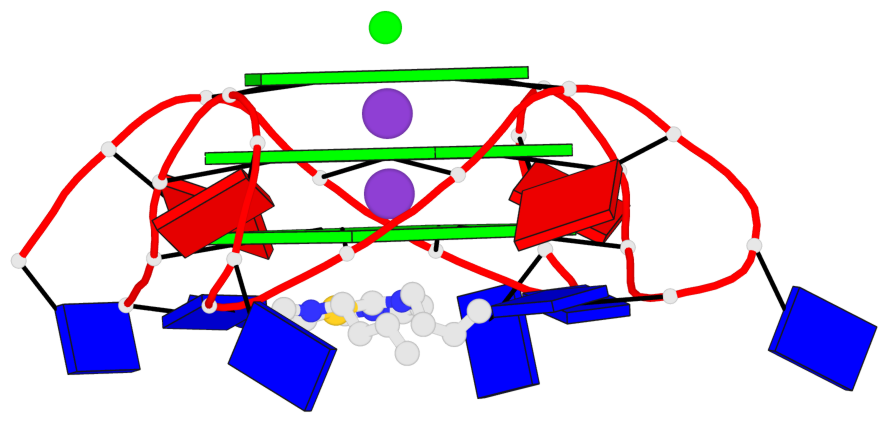
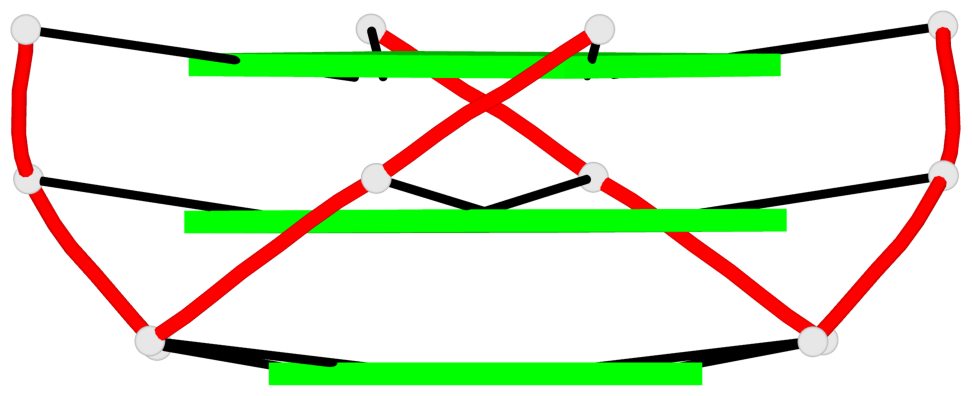
Base-block schematics in six views
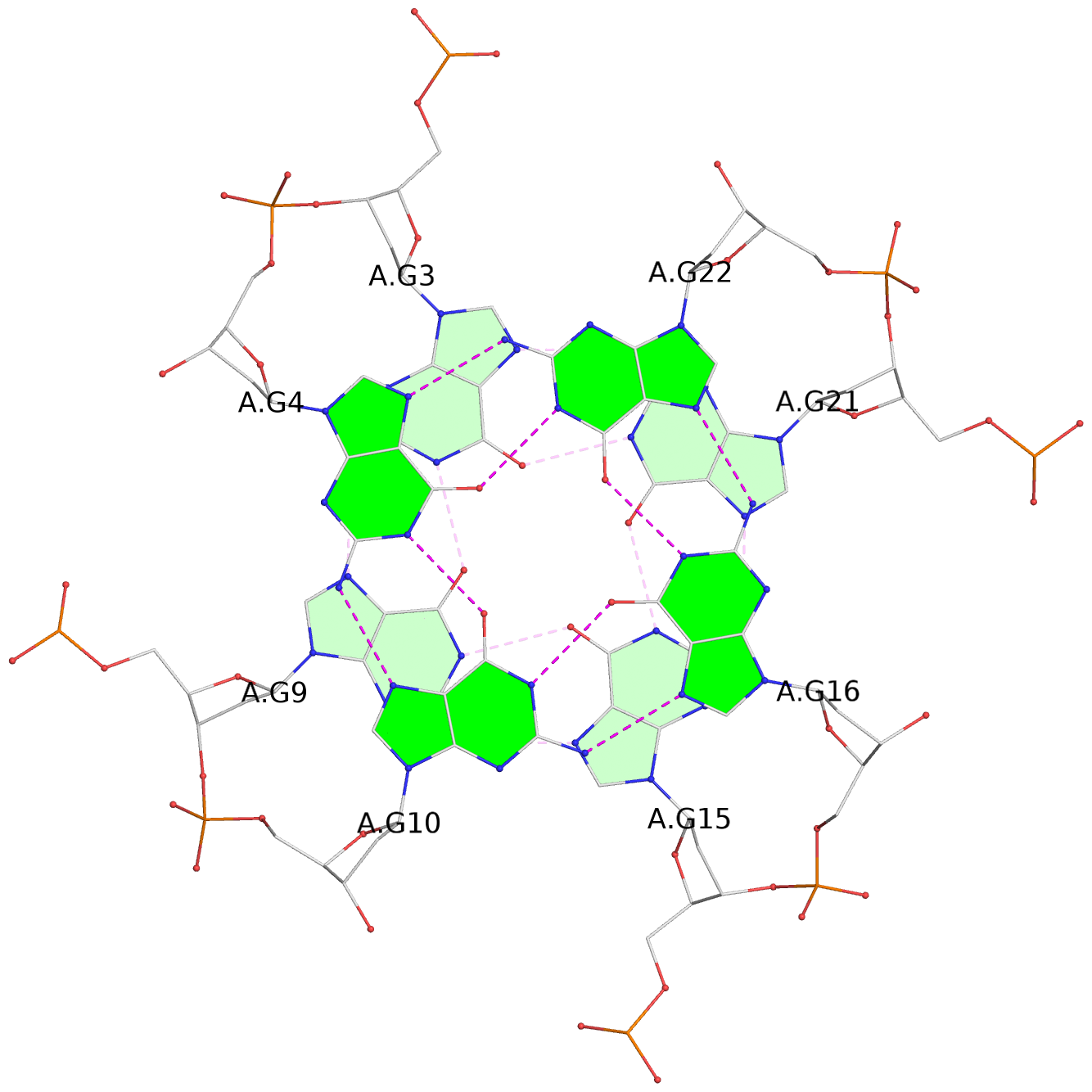
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.036 type=planar nts=4 GGGG A.DG3,A.DG9,A.DG15,A.DG21 2 glyco-bond=---- sugar=---- groove=---- planarity=0.031 type=planar nts=4 GGGG A.DG4,A.DG10,A.DG16,A.DG22 3 glyco-bond=---- sugar=---- groove=---- planarity=0.203 type=bowl nts=4 GGGG A.DG5,A.DG11,A.DG17,A.DG23
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.